import glob
import os
import requests
import s3fs
import fiona
import netCDF4 as nc
import h5netcdf
import xarray as xr
import pandas as pd
import geopandas as gpd
import numpy as np
import matplotlib.pyplot as plt
import hvplot.xarray
import earthaccess
from earthaccess import Auth, DataCollections, DataGranules, Store
from pathlib import Path
import zipfileFor an updated notebook using the latest data, see this notebook in the PO.DAAC Cookbook
SWOT Hydrology Science Application Tutorial on the Cloud
Retrieving SWOT attributes (WSE, width, slope) and plotting a longitudinal profile along a river or over a basin
Requirement
This tutorial can only be run in an AWS cloud instance running in us-west-2: NASA Earthdata Cloud data in S3 can be directly accessed via earthaccess python library; this access is limited to requests made within the US West (Oregon) (code: us-west-2) AWS region.
Earthdata Login
An Earthdata Login account is required to access data, as well as discover restricted data, from the NASA Earthdata system. Thus, to access NASA data, you need Earthdata Login. Please visit https://urs.earthdata.nasa.gov to register and manage your Earthdata Login account. This account is free to create and only takes a moment to set up.
This code runs using SWOT Level 2 Data Products (Version 1.1).
Notebook Authors: Arnaud Cerbelaud, Jeffrey Wade, NASA Jet Propulsion Laboratory - California Institute of Technology (Jan 2024)
Learning Objectives
- Retrieve SWOT hydrological attributes on river reaches within the AWS cloud (Cal/Val data). Query reaches by:
- River name
- Spatial bounding box
- Downstream tracing from reach id (e.g. headwater to outlet) for river longitudinal profiles
- Upstream tracing from reach id (e.g. outlet to full river network) for watershed analysis
- Plot a time series of WSE, width, slope data on the filtered data
- Visualize an interactive map of WSE, width, slope data on the filtered data
Last updated: 2 Feb 2024
Import Packages
Authenticate
Authenticate your Earthdata Login (EDL) information using the earthaccess python package as follows:
earthaccess.login() # Login with your EDL credentials if askedEnter your Earthdata Login username: cerbelaud
Enter your Earthdata password: ········<earthaccess.auth.Auth at 0x7f114ba7bd30>1. Retrieve SWOT hydrological attributes on river reaches within the AWS cloud (Cal/Val data)
What data should we download?
- Optional step: Get the .kmz file of SWOT passes/swaths (Version 1.1) and import it into Google Earth for visualization
- Determine which pass number corresponds to the river/basin you want to look at! #### Search for multiple days of data
# Enter pass number
pass_number = "009" # e.g. 009 for Connecticut in NA, 003 for Rhine in EU
# Enter continent code
continent_code = "NA" # e.g. "AF", "NA", "EU", "SI", "AS", "AU", "SA", "AR", "GR"
# Retrieves granule from the day we want, in this case by passing to `earthdata.search_data` function the data collection shortname and temporal bounds
river_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_RIVERSP_1.1',
temporal = ('2023-04-08 00:00:00', '2023-04-12 23:59:59'),
granule_name = "*Reach*_" + pass_number + "_" + continent_code + "*")
# Create fiona session to read data
fs_s3 = earthaccess.get_s3fs_session(results=river_results)
fiona_session=fiona.session.AWSSession(
aws_access_key_id=fs_s3.storage_options["key"],
aws_secret_access_key=fs_s3.storage_options["secret"],
aws_session_token=fs_s3.storage_options["token"]
)Granules found: 5Unzip selected files in Fiona session
# Initialize list of shapefiles containing all dates
SWOT_HR_shps = []
# Loop through queried granules to stack all acquisition dates
for j in range(len(river_results)):
# Get the link for each zip file
river_link = earthaccess.results.DataGranule.data_links(river_results[j], access='direct')[0]
# We use the zip+ prefix so fiona knows that we are operating on a zip file
river_shp_url = f"zip+{river_link}"
# Read shapefile
with fiona.Env(session=fiona_session):
SWOT_HR_shps.append(gpd.read_file(river_shp_url)) Aggregate unzipped files into dataframe
# Combine granules from all acquisition dates into one dataframe
SWOT_HR_df = gpd.GeoDataFrame(pd.concat(SWOT_HR_shps, ignore_index=True))
# Sort dataframe by reach_id and time
SWOT_HR_df = SWOT_HR_df.sort_values(['reach_id', 'time'])
SWOT_HR_df| reach_id | time | time_tai | time_str | p_lat | p_lon | river_name | wse | wse_u | wse_r_u | ... | p_wid_var | p_n_nodes | p_dist_out | p_length | p_maf | p_dam_id | p_n_ch_max | p_n_ch_mod | p_low_slp | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1958 | 72270400231 | -1.000000e+12 | -1.000000e+12 | no_data | 56.794630 | -66.461122 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 1618.079 | 53 | 230079.472 | 10630.774958 | -1.000000e+12 | 0 | 1 | 1 | 0 | LINESTRING (-66.48682 56.83442, -66.48645 56.8... |
| 2641 | 72270400231 | -1.000000e+12 | -1.000000e+12 | no_data | 56.794630 | -66.461122 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 1618.079 | 53 | 230079.472 | 10630.774958 | -1.000000e+12 | 0 | 1 | 1 | 0 | LINESTRING (-66.48682 56.83442, -66.48645 56.8... |
| 1959 | 72270400241 | -1.000000e+12 | -1.000000e+12 | no_data | 56.744361 | -66.341905 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 4831.128 | 53 | 240728.430 | 10648.958289 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-66.41602 56.75969, -66.41553 56.7... |
| 2642 | 72270400241 | -1.000000e+12 | -1.000000e+12 | no_data | 56.744361 | -66.341905 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 4831.128 | 53 | 240728.430 | 10648.958289 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-66.41602 56.75969, -66.41553 56.7... |
| 1960 | 72270400251 | 7.344991e+08 | 7.344991e+08 | 2023-04-11T03:31:02Z | 56.676087 | -66.219811 | no_data | 2.855340e+02 | 1.008930e+00 | 1.004910e+00 | ... | 412.975 | 58 | 252286.769 | 11558.339052 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-66.27812 56.71437, -66.27775 56.7... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 615 | 73130000061 | -1.000000e+12 | -1.000000e+12 | no_data | 41.424945 | -73.230960 | Housatonic River | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 10469.706 | 82 | 47161.191 | 16303.793847 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-73.17436 41.38295, -73.17465 41.3... |
| 1283 | 73130000061 | -1.000000e+12 | -1.000000e+12 | no_data | 41.424945 | -73.230960 | Housatonic River | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 10469.706 | 82 | 47161.191 | 16303.793847 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-73.17436 41.38295, -73.17465 41.3... |
| 1957 | 73130000061 | -1.000000e+12 | -1.000000e+12 | no_data | 41.424945 | -73.230960 | Housatonic River | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 10469.706 | 82 | 47161.191 | 16303.793847 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-73.17436 41.38295, -73.17465 41.3... |
| 2640 | 73130000061 | -1.000000e+12 | -1.000000e+12 | no_data | 41.424945 | -73.230960 | Housatonic River | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 10469.706 | 82 | 47161.191 | 16303.793847 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-73.17436 41.38295, -73.17465 41.3... |
| 3312 | 73130000061 | -1.000000e+12 | -1.000000e+12 | no_data | 41.424945 | -73.230960 | Housatonic River | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 10469.706 | 82 | 47161.191 | 16303.793847 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-73.17436 41.38295, -73.17465 41.3... |
3313 rows × 127 columns
Exploring the dataset
What acquisition dates and rivers do our downloaded files cover?
print('Available dates are:')
print(np.unique([i[:10] for i in SWOT_HR_df['time_str']]))
print('Available rivers are:')
print(np.unique([i for i in SWOT_HR_df['river_name']]))Available dates are:
['2023-04-08' '2023-04-09' '2023-04-10' '2023-04-11' '2023-04-12'
'no_data']
Available rivers are:
['Allagash River' 'Androscoggin River' 'Big Black River' 'Canal de fuite'
'Carrabassett River' 'Concord River' 'Concord River; Sudbury River'
'Connecticut River' 'Connecticut River; Westfield River'
'Connecticut River; White River' 'Dead River'
'Dead River (Kennebec River)' 'Deerfield River' 'Farmington River'
'Housatonic River' 'Ikarut River' 'Kennebec River' 'Komaktorvik River'
'Magalloway River' 'Merrimack River' 'Moose River'
'North Branch Penobscot River' 'Passumsic River' 'Pemigewasset River'
'Penobscot River West Branch' 'Quinebaug River' 'Saco River'
'Saguenay River' 'Saint Francis River' 'Saint John River'
'Saint Lawrence River' 'Sandy River' 'Shetucket River' 'Sudbury River'
'Thames River' 'West River' 'White River' 'no_data']Filter dataframe by river name of interest and plot selected reaches:
Note: Some rivers have multiple names, hence using the contains function
# Enter river name
river = "Connecticut River" # e.g. "Rhine", "Connecticut River"
## Filter dataframe
SWOT_HR_df_river = SWOT_HR_df[(SWOT_HR_df.river_name.str.contains(river))]
# Plot geopandas dataframe with 'explore' by reach id
SWOT_HR_df_river[['reach_id','river_name','geometry']].explore('reach_id', style_kwds=dict(weight=6))Filter dataframe by latitude/longitude of interest:
lat_start = {"Connecticut River": 41.5,
"Rhine": 47.5
}
lat_end = {"Connecticut River": 45,
"Rhine": 51
}
lon_start = {"Connecticut River": -74,
"Rhine": 7
}
lon_end = {"Connecticut River": -71,
"Rhine": 10
}
## Filter dataframe
SWOT_HR_df_box = SWOT_HR_df[(SWOT_HR_df.p_lat > lat_start[river]) & (SWOT_HR_df.p_lat < lat_end[river]) & (SWOT_HR_df.p_lon > lon_start[river]) & (SWOT_HR_df.p_lon < lon_end[river])]
# Plot geopandas dataframe with 'explore' by river name
SWOT_HR_df_box[['reach_id','river_name','geometry']].explore('river_name', style_kwds=dict(weight=6))2. River longitudinal profile: trace reaches downstream of given starting reach using rch_id_dn field
WARNING: This works as long as the data is exhaustive (no missing SWORD reaches)
First, let’s set up a dictionary relating all reaches in the dataset to their downstream neighbor
Note: rch_dn_dict[rch_id] gives a list of all the reaches directly downstream from rch_id
# Format rch_id_dn for dictionary. Rch_id_dn allows for multiple downstream reaches to be stored
# Also removes spaces in attribute field
rch_id_dn = [[x.strip() for x in SWOT_HR_df.rch_id_dn[j].split(',')] for j in range(0,len(SWOT_HR_df.rch_id_dn))]
# Filter upstream reach ids to remove 'no_data'
rch_id_dn_filter = [[x for x in dn_id if x.isnumeric()] for dn_id in rch_id_dn]
# Create lookup dictionary for river network topology: Downstream
rch_dn_dict = {SWOT_HR_df.reach_id[i]: rch_id_dn_filter[i] for i in range(len(SWOT_HR_df))}Then, starting from a given reach, let’s trace all connected downstream reaches
# Enter reach_id from which we will trace downstream (e.g. headwaters of the Connecticut River)
rch_dn_st = {"Connecticut River": '73120000691',
"Rhine": '23267000651'
}
# Initialize list to store downstream reaches, including starting reach
rch_dn_list = [rch_dn_st[river]]
# Retrieve first downstream id of starting reach and add to list
rch_dn_next = rch_dn_dict[rch_dn_st[river]][0]
# Trace next downstream reach until we hit the outlet (or here the last reach on file)
while len(rch_dn_next) != 0:
# Add reach to list if value exists
if len(rch_dn_next) != 0:
rch_dn_list.append(rch_dn_next)
# Recursively retrieve first downstream id of next reach
# Catch error if reach isn't in downloaded data
try:
rch_dn_next = rch_dn_dict[rch_dn_next][0]
except:
breakFinally, we filtered our downloaded data by the traced reaches to create a plot
# Filter downloaded data by downstream traced reaches
SWOT_dn_trace = SWOT_HR_df[SWOT_HR_df.reach_id.isin(rch_dn_list)]
# Remove reaches from rch_dn_list that are not present in SWOT data
rch_dn_list = [rch for rch in rch_dn_list if rch in SWOT_HR_df.reach_id.values]
SWOT_dn_trace[['reach_id','river_name','geometry']].explore('river_name', style_kwds=dict(weight=6))3. Watershed analysis: trace reaches upstream of starting reach using rch_id_up field
WARNING: This works as long as the data is exhaustive (no missing SWORD reaches)
First, let’s set up a dictionary relating all reaches in the dataset to their upstream neighbor
Note: rch_up_dict[rch_id] gives a list of all the reaches directly upstream from rch_id
# Format rch_id_up for dictionary. Rch_id_up allows for multiple upstream reaches to be stored
# Also removes spaces in attribute field
rch_id_up = [[x.strip() for x in SWOT_HR_df.rch_id_up[j].split(',')] for j in range(0,len(SWOT_HR_df.rch_id_up))]
# Filter upstream reach ids to remove 'no_data'
rch_id_up_fil = [[x for x in ups_id if x.isnumeric()] for ups_id in rch_id_up]
# Create lookup dictionary for river network topology: Upstream
rch_up_dict = {SWOT_HR_df.reach_id[i]: rch_id_up_fil[i] for i in range(len(SWOT_HR_df))}Then, starting from a given reach, let’s trace all connected upstream reaches
This adds a bit of complexity, as we need to keep track of multiple branches upstream of the starting reach.
# Enter reach_id from which we will trace upstream (e.g. outlet of the Connecticut River)
rch_up_st = {"Connecticut River": '73120000013',
"Rhine": '23265000051'
}
# Initialize list to store traced upstream reaches, including starting reach
rch_up_list = [rch_up_st[river]]
# Retrieve ids of reaches upstream of starting reach and add to list
rch_up_next = rch_up_dict[rch_up_st[river]]
# For upstream tracing, we need to set a list of next upstream ids to start while loop
rch_next_id = rch_up_next
# Loop until no more reaches to trace
while len(rch_next_id) != 0:
# Initialize list to store next upstream ids
rch_next_id = []
# Loop through next upstream ids for given reach
for rch_up_sel in rch_up_next:
# Get values of existing upstream ids of rch_up_next reaches
# If reach isn't in SWOT data (usually ghost reaches), continue to next reach
try:
rch_next_id = rch_next_id + rch_up_dict[rch_up_sel]
except:
continue
# Append id to list
rch_up_list.append(rch_up_sel)
# If reaches exist, add to list for next cycle of tracing
if len(rch_next_id) != 0:
rch_up_next = rch_next_idFinally, we filtered our downloaded data by the traced reaches to create a plot
# Filter downloaded data by upstream traced reaches
SWOT_up_trace = SWOT_HR_df[SWOT_HR_df.reach_id.isin(rch_up_list)]
# Remove reaches from rch_up_list that are not present in SWOT data
rch_up_list = [rch for rch in rch_up_list if rch in SWOT_HR_df.reach_id.values]
SWOT_up_trace[['reach_id','river_name','geometry']].explore('river_name', style_kwds=dict(weight=6))4. Visualize and plot a time series of WSE/width/slope longitudinal profiles
Let’s create a time series dataframe from the downstream filtered database
# Retrieve all possible acquisition dates (keeping only YYYY-MM-DD)
dates = np.unique([i[:10] for i in [x for x in SWOT_HR_df['time_str'] if x!='no_data']])
# Create a new database for time series analysis with unique reach_ids
SWOT_dn_trace_time = SWOT_dn_trace.set_index('reach_id').groupby(level=0) \
.apply(lambda df: df.reset_index(drop=True)) \
.unstack().sort_index(axis=1, level=1)
SWOT_dn_trace_time.columns = ['{}_{}'.format(x[0],dates[x[1]]) for x in SWOT_dn_trace_time.columns]Plot a longitudinal profile for selected SWOT variable
# Explore variables you could choose to plot
for var in ["wse","slope","width","len"]:
print(SWOT_dn_trace.columns[SWOT_dn_trace.columns.str.contains(var)])Index(['wse', 'wse_u', 'wse_r_u', 'wse_c', 'wse_c_u', 'area_wse', 'p_wse',
'p_wse_var'],
dtype='object')
Index(['slope', 'slope_u', 'slope_r_u', 'slope2', 'slope2_u', 'slope2_r_u'], dtype='object')
Index(['width', 'width_u', 'width_c', 'width_c_u', 'p_width'], dtype='object')
Index(['p_length'], dtype='object')# Enter variable of interest for plotting
varstr = "wse"# Find cumulative length on the longitudinal profile
length_list = np.nan_to_num([SWOT_dn_trace.p_length[SWOT_dn_trace.reach_id == rch].mean()/1000 for rch in rch_dn_list])
cumlength_list = np.cumsum(length_list)
## Plot a longitudinal profile from the downstream tracing database
## Plot a longitudinal profile from the downstream tracing database
plt.figure(figsize=(12,8))
for t in dates:
# Store the quantity of interest (wse, width etc.) at time t
value = SWOT_dn_trace_time.loc[rch_dn_list,varstr+'_'+t]
# Remove set negative values (bad observations) to NaN and forward fill NaNs
value[value < 0] = np.nan
value = value.ffill()
# Plot the data
plt.plot(cumlength_list, value, label = varstr+'_'+t)
plt.xlabel('Downstream Distance (km)')
plt.ylabel(varstr)
plt.legend()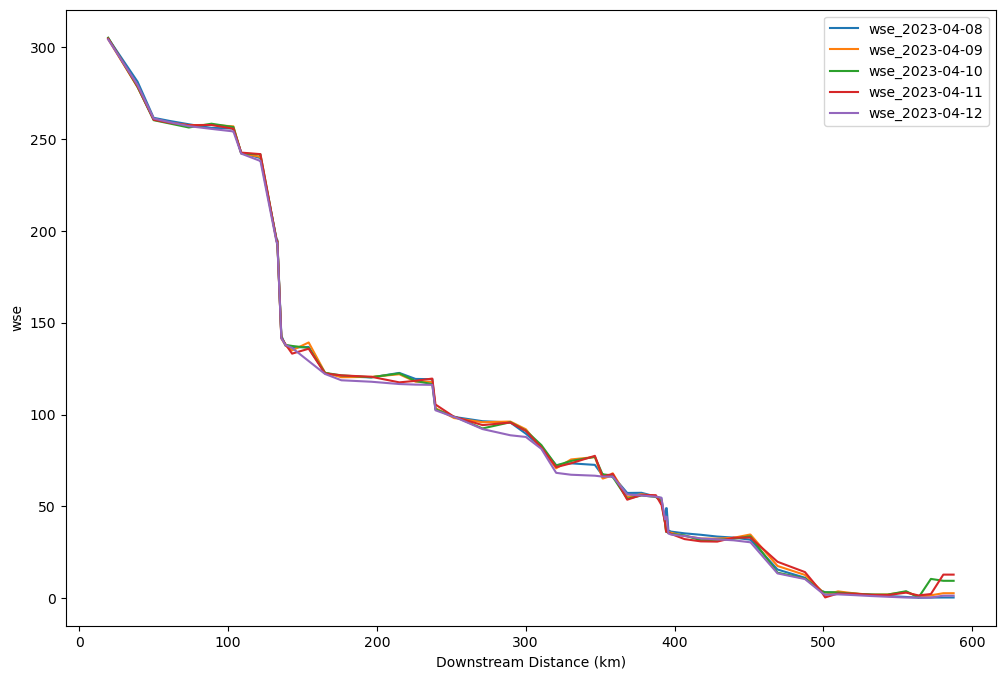
Map the longitudinal profile of selected SWOT variable
# Choose a date
date = dates[0]#Set one column as the active geometry in the new database
SWOT_dn_trace_time = SWOT_dn_trace_time.set_geometry("geometry_"+date)
#Set cleaner colorbar bounds for better visualization
vmin = np.percentile([i for i in SWOT_dn_trace_time[varstr+'_'+date] if i>0],5)
vmax = np.percentile([i for i in SWOT_dn_trace_time[varstr+'_'+date] if i>0],95)
# Interactive map
SWOT_dn_trace_time.explore(varstr+'_'+date,
vmin = vmin,
vmax = vmax,
cmap = "Blues", #"Blues",
control_scale = True,
tooltip = varstr+'_'+dates[0], # show "varstr+'_'+dates[0]" value in tooltip (on hover)
popup = True, # show all values in popup (on click)
#tiles = "CartoDB positron", # use "CartoDB positron" tiles
style_kwds=dict(weight=10)
)Supplemental
We can also map the upstream traced database
SWOT_up_trace_time = SWOT_up_trace.set_index('reach_id').groupby(level=0) \
.apply(lambda df: df.reset_index(drop=True)) \
.unstack().sort_index(axis=1, level=1)
SWOT_up_trace_time.columns = ['{}_{}'.format(x[0],dates[x[1]]) for x in SWOT_up_trace_time.columns]
SWOT_up_trace_time = SWOT_up_trace_time.set_geometry("geometry_"+date)
#Set cleaner colorbar bounds for better visualization
vmin = np.percentile([i for i in SWOT_up_trace_time[varstr+'_'+date] if i>0],5)
vmax = np.percentile([i for i in SWOT_up_trace_time[varstr+'_'+date] if i>0],95)
# Interactive map
SWOT_up_trace_time.explore(varstr+'_'+date,
vmin = vmin,
vmax = vmax,
cmap = "Blues", #"Blues",
control_scale = True,
tooltip = varstr+'_'+dates[0], # show "varstr+'_'+dates[0]" value in tooltip (on hover)
popup = True, # show all values in popup (on click)
tiles = "CartoDB positron", # use "CartoDB positron" tiles
style_kwds=dict(weight=5)
)How to filter out raster data to reveal water bodies?
Now you will also need the tile number (in addition to the pass number). You can find it on the .kmz file
# Enter tile number
tile_number = "116F"
# Retrieve granules from all days to find the cycle corresponding to the desired date
raster_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_Raster_1.1',
temporal = ('2023-04-01 00:00:00', '2023-04-22 23:59:59'),
granule_name = "*100m*_" + pass_number + "_" + tile_number + "*")
# here we filter by files with '100m' in the name, pass=009, scene = 116F (tile = 232L) for Connecticut
# pass=003, scene = 120F (tile = 239R) for the RhineGranules found: 15Display details of downloaded files
print([i for i in raster_results])[Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 484, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-08T03:55:38.288Z', 'BeginningDateTime': '2023-04-08T03:55:18.398Z'}}
Size(MB): 70.40732002258301
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_484_009_116F_20230408T035517_20230408T035538_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 485, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-09T03:46:16.448Z', 'BeginningDateTime': '2023-04-09T03:45:56.558Z'}}
Size(MB): 68.41873073577881
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_485_009_116F_20230409T034556_20230409T034616_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 486, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-10T03:36:54.573Z', 'BeginningDateTime': '2023-04-10T03:36:34.684Z'}}
Size(MB): 68.47044467926025
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_486_009_116F_20230410T033634_20230410T033655_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 487, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-11T03:27:32.662Z', 'BeginningDateTime': '2023-04-11T03:27:12.772Z'}}
Size(MB): 69.02541446685791
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_487_009_116F_20230411T032712_20230411T032733_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 488, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-12T03:18:10.720Z', 'BeginningDateTime': '2023-04-12T03:17:50.831Z'}}
Size(MB): 67.56003761291504
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_488_009_116F_20230412T031750_20230412T031811_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 489, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-13T03:08:48.751Z', 'BeginningDateTime': '2023-04-13T03:08:28.862Z'}}
Size(MB): 68.72685241699219
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_489_009_116F_20230413T030828_20230413T030849_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.4, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 490, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-14T02:59:26.753Z', 'BeginningDateTime': '2023-04-14T02:59:06.866Z'}}
Size(MB): 66.47898387908936
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_490_009_116F_20230414T025906_20230414T025927_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 491, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-15T02:50:04.726Z', 'BeginningDateTime': '2023-04-15T02:49:44.836Z'}}
Size(MB): 67.29588985443115
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_491_009_116F_20230415T024944_20230415T025005_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 492, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-16T02:40:43.207Z', 'BeginningDateTime': '2023-04-16T02:40:22.244Z'}}
Size(MB): 0.933258056640625
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_492_009_116F_20230416T024022_20230416T024043_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 493, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-17T02:31:21.112Z', 'BeginningDateTime': '2023-04-17T02:31:00.151Z'}}
Size(MB): 0.9332094192504883
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_493_009_116F_20230417T023100_20230417T023121_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 494, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-18T02:21:58.990Z', 'BeginningDateTime': '2023-04-18T02:21:38.024Z'}}
Size(MB): 0.9332304000854492
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_494_009_116F_20230418T022138_20230418T022158_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 495, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-19T02:12:36.279Z', 'BeginningDateTime': '2023-04-19T02:12:16.394Z'}}
Size(MB): 63.773990631103516
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_495_009_116F_20230419T021215_20230419T021236_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 496, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-20T02:03:14.075Z', 'BeginningDateTime': '2023-04-20T02:02:54.187Z'}}
Size(MB): 62.185853004455566
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_496_009_116F_20230420T020253_20230420T020314_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 497, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-21T01:53:51.831Z', 'BeginningDateTime': '2023-04-21T01:53:31.942Z'}}
Size(MB): 61.36593437194824
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_497_009_116F_20230421T015331_20230421T015352_PIB0_01.nc'], Collection: {'Version': '1.1', 'ShortName': 'SWOT_L2_HR_Raster_1.1'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'StartLatitude': -77.66, 'EndLatitude': 77.66, 'AscendingCrossing': -80.41, 'StartDirection': 'A', 'EndDirection': 'A'}, 'Track': {'Cycle': 498, 'Passes': [{'Pass': 9, 'Tiles': ['230L', '231L', '232L', '233L', '230R', '231R', '232R', '233R']}]}}}
Temporal coverage: {'RangeDateTime': {'EndingDateTime': '2023-04-22T01:44:29.616Z', 'BeginningDateTime': '2023-04-22T01:44:09.731Z'}}
Size(MB): 61.341376304626465
Data: ['https://archive.swot.podaac.earthdata.nasa.gov/podaac-swot-ops-cumulus-protected/SWOT_L2_HR_Raster_1.1/SWOT_L2_HR_Raster_100m_UTM19T_N_x_x_x_498_009_116F_20230422T014409_20230422T014430_PIB0_01.nc']]Select a single file for visualization
# Let's look at cycle 485, on the second day of calval
# Enter cycle
cycle = "485"
raster_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_Raster_1.1',
temporal = ('2023-04-01 00:00:00', '2023-04-22 23:59:59'),
granule_name = "*100m*" + cycle + "_" + pass_number + "_" + tile_number + "*")
# here we filter by files with '100m' in the name, cycle=484, pass=009, scene = 116F for Connecticut
# cycle=485, pass=003, scene = 120F for the RhineGranules found: 1Open file using fiona AWS session and read file using xarray
fs_s3 = earthaccess.get_s3fs_session(results=raster_results)
# get link for file and open it
raster_link = earthaccess.results.DataGranule.data_links(raster_results[0], access='direct')[0]
s3_file_obj5 = fs_s3.open(raster_link, mode='rb')
# Open data with xarray
ds_raster = xr.open_dataset(s3_file_obj5, engine='h5netcdf')Plot raster data using custom filters to improve image quality
# Plot the data with a filter on water_frac > x, or sig0>50
ds_raster.wse.where(ds_raster.water_frac > 0.5).hvplot.image(y='y', x='x').opts(cmap='Blues', clim=(vmin,vmax), width=1000, height=750)