# Core libraries for this tutorial
# Available via `pip install zarr zarr-eosdis-store`
from eosdis_store import EosdisStore
import xarray as xr
# Other Python libraries
import requests
from pqdm.threads import pqdm
from matplotlib import animation, pyplot as plt
from IPython.core.display import display, HTML
# Python standard library imports
from pprint import pprintZarr Access for NetCDF4 files
imported on: 2026-02-13
This notebook is from NASA Openscapes 2021 Cloud Hackathon Repository
The original source for this document is https://github.com/NASA-Openscapes/2021-Cloud-Hackathon/blob/main/tutorials/09_Zarr_Access.ipynb
09. Zarr Access for NetCDF4 files
Timing:
- Exercise: 45 minutes
Summary
Zarr is an open source library for storing N-dimensional array data. It supports multidimensional arrays with attributes and dimensions similar to NetCDF4, and it can be read by XArray. Zarr is often used for data held in cloud object storage (like Amazon S3), because it is better optimized for these situations than NetCDF4.
The zarr-eosdis-store library allows NASA EOSDIS NetCDF4 files to be read more efficiently by transferring only file metadata and data needed for computation in a small number of requests, rather than moving the whole file or making many small requests. It works by making the files directly readable by the Zarr Python library and XArray across a network. To use it, files must have a corresponding metadata file ending in .dmrpp, which increasingly true for cloud-accessible EOSDIS data. https://github.com/nasa/zarr-eosdis-store
The zarr-eosdis-store library provides several benefits over downloading EOSDIS data files and accessing them using XArray, NetCDF4, or HDF5 Python libraries:
- It only downloads the chunks of data you actually read, so if you don’t read all variables or the full spatiotemporal extent of a file, you usually won’t spend time downloading those portions of the file
- It parallelizes and optimizes downloads for the portions of files you do read, so download speeds can be faster in general
- It automatically interoperates with Earthdata Login if you have a .netrc file set up
- It is aware of some EOSDIS cloud implementation quirks and provides caching that can save time for repeated requests to individual files
It can also be faster than using XArray pointing NetCDF4 files with s3:// URLs, depending on the file’s internal structure, and is often more convenient.
Consider using this library when: 1. The portion of the data file you need to use is much smaller than the full file, e.g. in cases of spatial subsets or reading a single variable from a file containing several 1. s3:// URLs are not readily available 1. Code need to run outside of the AWS cloud or us-west-2 region or in a hybrid cloud / non-cloud manner 1. s3:// access using XArray seems slower than you would expect (possibly due to unoptimized internal file structure) 1. No readily-available, public, cloud-optimized version of the data exists already. The example we show is also available as an AWS Public Dataset: https://registry.opendata.aws/mur/ 1. Adding “.dmrpp” to the end of a data URL returns a file
Objectives
- Build on prior knowledge from CMR and Earthdata Login tutorials
- Work through an example of using the EOSDIS Zarr Store to access data using XArray
- Learn about the Zarr format and library for accessing data in the cloud
Exercise
In this exercise, we will be using the eosdis-zarr-store library to aggregate and analyze a month of sea surface temperature for the Great Lakes region
Set up
Import Required Packages
Also set the width / height for plots we show
plt.rcParams['figure.figsize'] = 12, 6Set Dataset, Time, and Region of Interest
Look in PO.DAAC’s cloud archive for Group for High Resolution Sea Surface Temperature (GHRSST) Level 4 Multiscale Ultrahigh Resolution (MUR) data
data_provider = 'POCLOUD'
mur_short_name = 'MUR-JPL-L4-GLOB-v4.1'Looking for data from the month of September over the Great Lakes
start_time = '2021-09-01T21:00:00Z'
end_time = '2021-09-30T20:59:59Z'
# Bounding box around the Great Lakes
lats = slice(41, 49)
lons = slice(-93, -76)
# Some other possibly interesting bounding boxes:
# Hawaiian Islands
# lats = slice(18, 22.5)
# lons = slice(-161, -154)
# Mediterranean Sea
# lats = slice(29, 45)
# lons = slice(-7, 37)Find URLs for the dataset and AOI
Set up a CMR granules search for our area of interest, as we saw in prior tutorials
cmr_url = 'https://cmr.earthdata.nasa.gov/search/granules.json'Search for granules in our area of interest, expecting one granule per day of September
response = requests.get(cmr_url,
params={
'provider': data_provider,
'short_name': mur_short_name,
'temporal': f'{start_time},{end_time}',
'bounding_box': f'{lons.start},{lats.start},{lons.stop},{lats.stop}',
'page_size': 2000,
}
)granules = response.json()['feed']['entry']
for granule in granules:
print(granule['title'])20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210902090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210903090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210904090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210905090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210906090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210907090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210908090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210909090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210910090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210911090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210912090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210913090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210914090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210915090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210916090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210917090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210918090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210919090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210920090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210921090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210922090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210923090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210924090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210925090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210926090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210927090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210928090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210929090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1
20210930090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1pprint(granules[0]){'boxes': ['-90 -180 90 180'],
'browse_flag': False,
'collection_concept_id': 'C1996881146-POCLOUD',
'coordinate_system': 'CARTESIAN',
'data_center': 'POCLOUD',
'dataset_id': 'GHRSST Level 4 MUR Global Foundation Sea Surface Temperature '
'Analysis (v4.1)',
'day_night_flag': 'UNSPECIFIED',
'granule_size': '9.059906005859375E-5',
'id': 'G2113241213-POCLOUD',
'links': [{'href': 's3://podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'hreflang': 'en-US',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/s3#',
'title': 'This link provides direct download access via S3 to the '
'granule.'},
{'href': 'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'hreflang': 'en-US',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/data#',
'title': 'Download '
'20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc'},
{'href': 'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-public/MUR-JPL-L4-GLOB-v4.1/20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc.md5',
'hreflang': 'en-US',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/metadata#',
'title': 'Download '
'20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc.md5'},
{'href': 'https://archive.podaac.earthdata.nasa.gov/s3credentials',
'hreflang': 'en-US',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/metadata#',
'title': 'api endpoint to retrieve temporary credentials valid for '
'same-region direct s3 access'},
{'href': 'https://opendap.earthdata.nasa.gov/collections/C1996881146-POCLOUD/granules/20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1',
'hreflang': 'en-US',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/service#',
'title': 'OPeNDAP request URL'},
{'href': 'https://github.com/nasa/podaac_tools_and_services/tree/master/subset_opendap',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://ghrsst.jpl.nasa.gov',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://earthdata.nasa.gov/esds/competitive-programs/measures/mur-sst',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/metadata#'},
{'href': 'http://journals.ametsoc.org/doi/abs/10.1175/1520-0426%281998%29015%3C0741:BSHWSS%3E2.0.CO;2',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://podaac-tools.jpl.nasa.gov/drive/files/OceanTemperature/ghrsst/docs/GDS20r5.pdf',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://github.com/podaac/data-readers',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://doi.org/10.1016/j.rse.2017.07.029',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://registry.opendata.aws/mur/#usageexa',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/metadata#'},
{'href': 'http://www.ghrsst.org',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://podaac.jpl.nasa.gov/CitingPODAAC',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://cmr.earthdata.nasa.gov/virtual-directory/collections/C1996881146-POCLOUD ',
'hreflang': 'en-US',
'inherited': True,
'length': '300.0MB',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/data#'},
{'href': ' '
'https://search.earthdata.nasa.gov/search/granules?p=C1996881146-POCLOUD ',
'hreflang': 'en-US',
'inherited': True,
'length': '700.0MB',
'rel': 'http://esipfed.org/ns/fedsearch/1.1/data#'},
{'href': 'https://podaac.jpl.nasa.gov/MEaSUREs-MUR',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'},
{'href': 'https://github.com/podaac/tutorials/blob/master/notebooks/SWOT-EA-2021/Colocate_satellite_insitu_ocean.ipynb',
'hreflang': 'en-US',
'inherited': True,
'rel': 'http://esipfed.org/ns/fedsearch/1.1/documentation#'}],
'online_access_flag': True,
'original_format': 'UMM_JSON',
'time_end': '2021-09-01T21:00:00.000Z',
'time_start': '2021-08-31T21:00:00.000Z',
'title': '20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1',
'updated': '2021-09-10T07:29:40.511Z'}urls = []
for granule in granules:
for link in granule['links']:
if link['rel'].endswith('/data#'):
urls.append(link['href'])
break
pprint(urls)['https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210901090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210902090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210903090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210904090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210905090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210906090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210907090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210908090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210909090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210910090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210911090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210912090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210913090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210914090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210915090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210916090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210917090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210918090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210919090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210920090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210921090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210922090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210923090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210924090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210925090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210926090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210927090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210928090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210929090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc',
'https://archive.podaac.earthdata.nasa.gov/podaac-ops-cumulus-protected/MUR-JPL-L4-GLOB-v4.1/20210930090000-JPL-L4_GHRSST-SSTfnd-MUR-GLOB-v02.0-fv04.1.nc']Open and view our AOI without downloading a whole file
Check to see if we can use an efficient partial-access technique
response = requests.head(f'{urls[0]}.dmrpp')
print('Can we use EosdisZarrStore and XArray to access these files more efficiently?')
print('Yes' if response.ok else 'No')Can we use EosdisZarrStore and XArray to access these files more efficiently?
YesOpen our first URL using the Zarr library
url = urls[0]
ds = xr.open_zarr(EosdisStore(url), consolidated=False)That’s it! No downloads, temporary credentials, or S3 filesystems. Hereafter, we interact with the ds variable as with any XArray dataset. We need not worry about the EosdisStore anymore.
View the file’s variable structure
ds<xarray.Dataset>
Dimensions: (time: 1, lat: 17999, lon: 36000)
Coordinates:
* lat (lat) float32 -89.99 -89.98 -89.97 ... 89.97 89.98 89.99
* lon (lon) float32 -180.0 -180.0 -180.0 ... 180.0 180.0 180.0
* time (time) datetime64[ns] 2021-09-01T09:00:00
Data variables:
analysed_sst (time, lat, lon) float32 dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
analysis_error (time, lat, lon) float32 dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
dt_1km_data (time, lat, lon) timedelta64[ns] dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
mask (time, lat, lon) float32 dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
sea_ice_fraction (time, lat, lon) float32 dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
sst_anomaly (time, lat, lon) float32 dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid- time: 1
- lat: 17999
- lon: 36000
- lat(lat)float32-89.99 -89.98 ... 89.98 89.99
- long_name :
- latitude
- standard_name :
- latitude
- axis :
- Y
- units :
- degrees_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- comment :
- geolocations inherited from the input data without correction
array([-89.99, -89.98, -89.97, ..., 89.97, 89.98, 89.99], dtype=float32)
- lon(lon)float32-180.0 -180.0 ... 180.0 180.0
- long_name :
- longitude
- standard_name :
- longitude
- axis :
- X
- units :
- degrees_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- comment :
- geolocations inherited from the input data without correction
array([-179.99, -179.98, -179.97, ..., 179.98, 179.99, 180. ], dtype=float32) - time(time)datetime64[ns]2021-09-01T09:00:00
- long_name :
- reference time of sst field
- standard_name :
- time
- axis :
- T
- comment :
- Nominal time of analyzed fields
array(['2021-09-01T09:00:00.000000000'], dtype='datetime64[ns]')
- analysed_sst(time, lat, lon)float32dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
- long_name :
- analysed sea surface temperature
- standard_name :
- sea_surface_foundation_temperature
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- \"Final\" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
Array Chunk Bytes 2.41 GiB 7.99 MiB Shape (1, 17999, 36000) (1, 1023, 2047) Count 325 Tasks 324 Chunks Type float32 numpy.ndarray - analysis_error(time, lat, lon)float32dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
- long_name :
- estimated error standard deviation of analysed_sst
- units :
- kelvin
- valid_min :
- 0
- valid_max :
- 32767
- comment :
- uncertainty in \"analysed_sst\"
Array Chunk Bytes 2.41 GiB 7.99 MiB Shape (1, 17999, 36000) (1, 1023, 2047) Count 325 Tasks 324 Chunks Type float32 numpy.ndarray - dt_1km_data(time, lat, lon)timedelta64[ns]dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
- long_name :
- time to most recent 1km data
- valid_min :
- -127
- valid_max :
- 127
- source :
- MODIS and VIIRS pixels ingested by MUR
- comment :
- The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. \"Fill value\" indicates absence of such 1km data at the grid point.
Array Chunk Bytes 4.83 GiB 31.96 MiB Shape (1, 17999, 36000) (1, 1447, 2895) Count 170 Tasks 169 Chunks Type timedelta64[ns] numpy.ndarray - mask(time, lat, lon)float32dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
- long_name :
- sea/land field composite mask
- valid_min :
- 1
- valid_max :
- 31
- flag_masks :
- [1, 2, 4, 8, 16]
- flag_meanings :
- open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid
- comment :
- mask can be used to further filter the data.
- source :
- GMT \"grdlandmask\", ice flag from sea_ice_fraction data
Array Chunk Bytes 2.41 GiB 15.98 MiB Shape (1, 17999, 36000) (1, 1447, 2895) Count 170 Tasks 169 Chunks Type float32 numpy.ndarray - sea_ice_fraction(time, lat, lon)float32dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
- long_name :
- sea ice area fraction
- standard_name :
- sea_ice_area_fraction
- valid_min :
- 0
- valid_max :
- 100
- source :
- EUMETSAT OSI-SAF, copyright EUMETSAT
- comment :
- ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach.
Array Chunk Bytes 2.41 GiB 15.98 MiB Shape (1, 17999, 36000) (1, 1447, 2895) Count 170 Tasks 169 Chunks Type float32 numpy.ndarray - sst_anomaly(time, lat, lon)float32dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
- long_name :
- SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- anomaly reference to the day-of-year average between 2003 and 2014
Array Chunk Bytes 2.41 GiB 7.99 MiB Shape (1, 17999, 36000) (1, 1023, 2047) Count 325 Tasks 324 Chunks Type float32 numpy.ndarray
- Conventions :
- CF-1.7
- title :
- Daily MUR SST, Final product
- summary :
- A merged, multi-sensor L4 Foundation SST analysis product from JPL.
- references :
- http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST
- institution :
- Jet Propulsion Laboratory
- history :
- created at nominal 4-day latency; replaced nrt (1-day latency) version.
- comment :
- MUR = \"Multi-scale Ultra-high Resolution\"
- license :
- These data are available free of charge under data policy of JPL PO.DAAC.
- id :
- MUR-JPL-L4-GLOB-v04.1
- naming_authority :
- org.ghrsst
- product_version :
- 04.1
- uuid :
- 27665bc0-d5fc-11e1-9b23-0800200c9a66
- gds_version_id :
- 2.0
- netcdf_version_id :
- 4.1
- date_created :
- 20210910T072132Z
- start_time :
- 20210901T090000Z
- stop_time :
- 20210901T090000Z
- time_coverage_start :
- 20210831T210000Z
- time_coverage_end :
- 20210901T210000Z
- file_quality_level :
- 3
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
- platform :
- Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships
- sensor :
- MODIS, AMSR2, AVHRR, in-situ
- Metadata_Conventions :
- Unidata Observation Dataset v1.0
- metadata_link :
- http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1
- keywords :
- Oceans > Ocean Temperature > Sea Surface Temperature
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- southernmost_latitude :
- -90.0
- northernmost_latitude :
- 90.0
- westernmost_longitude :
- -180.0
- easternmost_longitude :
- 180.0
- spatial_resolution :
- 0.01 degrees
- geospatial_lat_units :
- degrees north
- geospatial_lat_resolution :
- 0.009999999776
- geospatial_lon_units :
- degrees east
- geospatial_lon_resolution :
- 0.009999999776
- acknowledgment :
- Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program.
- creator_name :
- JPL MUR SST project
- creator_email :
- ghrsst@podaac.jpl.nasa.gov
- creator_url :
- http://mur.jpl.nasa.gov
- project :
- NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program
- publisher_name :
- GHRSST Project Office
- publisher_url :
- http://www.ghrsst.org
- publisher_email :
- ghrsst-po@nceo.ac.uk
- processing_level :
- L4
- cdm_data_type :
- grid
ds.analysed_sst<xarray.DataArray 'analysed_sst' (time: 1, lat: 17999, lon: 36000)>
dask.array<open_dataset-4d5a9a1e1fda090e80524b67b2e413c6analysed_sst, shape=(1, 17999, 36000), dtype=float32, chunksize=(1, 1023, 2047), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 -89.99 -89.98 -89.97 -89.96 ... 89.97 89.98 89.99
* lon (lon) float32 -180.0 -180.0 -180.0 -180.0 ... 180.0 180.0 180.0
* time (time) datetime64[ns] 2021-09-01T09:00:00
Attributes:
long_name: analysed sea surface temperature
standard_name: sea_surface_foundation_temperature
units: kelvin
valid_min: -32767
valid_max: 32767
comment: \"Final\" version using Multi-Resolution Variational Anal...
source: MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, A...- time: 1
- lat: 17999
- lon: 36000
- dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
Array Chunk Bytes 2.41 GiB 7.99 MiB Shape (1, 17999, 36000) (1, 1023, 2047) Count 325 Tasks 324 Chunks Type float32 numpy.ndarray - lat(lat)float32-89.99 -89.98 ... 89.98 89.99
- long_name :
- latitude
- standard_name :
- latitude
- axis :
- Y
- units :
- degrees_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- comment :
- geolocations inherited from the input data without correction
array([-89.99, -89.98, -89.97, ..., 89.97, 89.98, 89.99], dtype=float32)
- lon(lon)float32-180.0 -180.0 ... 180.0 180.0
- long_name :
- longitude
- standard_name :
- longitude
- axis :
- X
- units :
- degrees_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- comment :
- geolocations inherited from the input data without correction
array([-179.99, -179.98, -179.97, ..., 179.98, 179.99, 180. ], dtype=float32) - time(time)datetime64[ns]2021-09-01T09:00:00
- long_name :
- reference time of sst field
- standard_name :
- time
- axis :
- T
- comment :
- Nominal time of analyzed fields
array(['2021-09-01T09:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- analysed sea surface temperature
- standard_name :
- sea_surface_foundation_temperature
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- \"Final\" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
sst = ds.analysed_sst.sel(lat=lats, lon=lons)
sst<xarray.DataArray 'analysed_sst' (time: 1, lat: 801, lon: 1701)>
dask.array<getitem, shape=(1, 801, 1701), dtype=float32, chunksize=(1, 601, 1536), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 41.0 41.01 41.02 41.03 ... 48.97 48.98 48.99 49.0
* lon (lon) float32 -93.0 -92.99 -92.98 -92.97 ... -76.02 -76.01 -76.0
* time (time) datetime64[ns] 2021-09-01T09:00:00
Attributes:
long_name: analysed sea surface temperature
standard_name: sea_surface_foundation_temperature
units: kelvin
valid_min: -32767
valid_max: 32767
comment: \"Final\" version using Multi-Resolution Variational Anal...
source: MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, A...- time: 1
- lat: 801
- lon: 1701
- dask.array<chunksize=(1, 200, 1536), meta=np.ndarray>
Array Chunk Bytes 5.20 MiB 3.52 MiB Shape (1, 801, 1701) (1, 601, 1536) Count 329 Tasks 4 Chunks Type float32 numpy.ndarray - lat(lat)float3241.0 41.01 41.02 ... 48.99 49.0
- long_name :
- latitude
- standard_name :
- latitude
- axis :
- Y
- units :
- degrees_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- comment :
- geolocations inherited from the input data without correction
array([41. , 41.01, 41.02, ..., 48.98, 48.99, 49. ], dtype=float32)
- lon(lon)float32-93.0 -92.99 ... -76.01 -76.0
- long_name :
- longitude
- standard_name :
- longitude
- axis :
- X
- units :
- degrees_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- comment :
- geolocations inherited from the input data without correction
array([-93. , -92.99, -92.98, ..., -76.02, -76.01, -76. ], dtype=float32)
- time(time)datetime64[ns]2021-09-01T09:00:00
- long_name :
- reference time of sst field
- standard_name :
- time
- axis :
- T
- comment :
- Nominal time of analyzed fields
array(['2021-09-01T09:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- analysed sea surface temperature
- standard_name :
- sea_surface_foundation_temperature
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- \"Final\" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
sst.plot()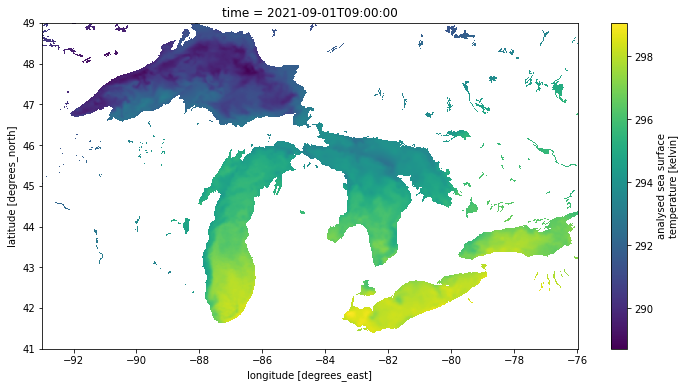
Aggregate and analyze 30 files
Set up a function to open all of our URLs as XArrays in parallel
def open_as_zarr_xarray(url):
return xr.open_zarr(EosdisStore(url), consolidated=False)
datasets = pqdm(urls, open_as_zarr_xarray, n_jobs=30)Combine the individual file-based datasets into a single xarray dataset with a time axis
ds = xr.concat(datasets, 'time')
ds<xarray.Dataset>
Dimensions: (time: 30, lat: 17999, lon: 36000)
Coordinates:
* lat (lat) float32 -89.99 -89.98 -89.97 ... 89.97 89.98 89.99
* lon (lon) float32 -180.0 -180.0 -180.0 ... 180.0 180.0 180.0
* time (time) datetime64[ns] 2021-09-01T09:00:00 ... 2021-09-3...
Data variables:
analysed_sst (time, lat, lon) float32 dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
analysis_error (time, lat, lon) float32 dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
dt_1km_data (time, lat, lon) timedelta64[ns] dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
mask (time, lat, lon) float32 dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
sea_ice_fraction (time, lat, lon) float32 dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
sst_anomaly (time, lat, lon) float32 dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7
title: Daily MUR SST, Final product
summary: A merged, multi-sensor L4 Foundation SST anal...
references: http://podaac.jpl.nasa.gov/Multi-scale_Ultra-...
institution: Jet Propulsion Laboratory
history: created at nominal 4-day latency; replaced nr...
... ...
project: NASA Making Earth Science Data Records for Us...
publisher_name: GHRSST Project Office
publisher_url: http://www.ghrsst.org
publisher_email: ghrsst-po@nceo.ac.uk
processing_level: L4
cdm_data_type: grid- time: 30
- lat: 17999
- lon: 36000
- lat(lat)float32-89.99 -89.98 ... 89.98 89.99
- long_name :
- latitude
- standard_name :
- latitude
- axis :
- Y
- units :
- degrees_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- comment :
- geolocations inherited from the input data without correction
array([-89.99, -89.98, -89.97, ..., 89.97, 89.98, 89.99], dtype=float32)
- lon(lon)float32-180.0 -180.0 ... 180.0 180.0
- long_name :
- longitude
- standard_name :
- longitude
- axis :
- X
- units :
- degrees_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- comment :
- geolocations inherited from the input data without correction
array([-179.99, -179.98, -179.97, ..., 179.98, 179.99, 180. ], dtype=float32) - time(time)datetime64[ns]2021-09-01T09:00:00 ... 2021-09-...
- long_name :
- reference time of sst field
- standard_name :
- time
- axis :
- T
- comment :
- Nominal time of analyzed fields
array(['2021-09-01T09:00:00.000000000', '2021-09-02T09:00:00.000000000', '2021-09-03T09:00:00.000000000', '2021-09-04T09:00:00.000000000', '2021-09-05T09:00:00.000000000', '2021-09-06T09:00:00.000000000', '2021-09-07T09:00:00.000000000', '2021-09-08T09:00:00.000000000', '2021-09-09T09:00:00.000000000', '2021-09-10T09:00:00.000000000', '2021-09-11T09:00:00.000000000', '2021-09-12T09:00:00.000000000', '2021-09-13T09:00:00.000000000', '2021-09-14T09:00:00.000000000', '2021-09-15T09:00:00.000000000', '2021-09-16T09:00:00.000000000', '2021-09-17T09:00:00.000000000', '2021-09-18T09:00:00.000000000', '2021-09-19T09:00:00.000000000', '2021-09-20T09:00:00.000000000', '2021-09-21T09:00:00.000000000', '2021-09-22T09:00:00.000000000', '2021-09-23T09:00:00.000000000', '2021-09-24T09:00:00.000000000', '2021-09-25T09:00:00.000000000', '2021-09-26T09:00:00.000000000', '2021-09-27T09:00:00.000000000', '2021-09-28T09:00:00.000000000', '2021-09-29T09:00:00.000000000', '2021-09-30T09:00:00.000000000'], dtype='datetime64[ns]')
- analysed_sst(time, lat, lon)float32dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
- long_name :
- analysed sea surface temperature
- standard_name :
- sea_surface_foundation_temperature
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- \"Final\" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
Array Chunk Bytes 72.42 GiB 7.99 MiB Shape (30, 17999, 36000) (1, 1023, 2047) Count 19470 Tasks 9720 Chunks Type float32 numpy.ndarray - analysis_error(time, lat, lon)float32dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
- long_name :
- estimated error standard deviation of analysed_sst
- units :
- kelvin
- valid_min :
- 0
- valid_max :
- 32767
- comment :
- uncertainty in \"analysed_sst\"
Array Chunk Bytes 72.42 GiB 7.99 MiB Shape (30, 17999, 36000) (1, 1023, 2047) Count 19470 Tasks 9720 Chunks Type float32 numpy.ndarray - dt_1km_data(time, lat, lon)timedelta64[ns]dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
- long_name :
- time to most recent 1km data
- valid_min :
- -127
- valid_max :
- 127
- source :
- MODIS and VIIRS pixels ingested by MUR
- comment :
- The grid value is hours between the analysis time and the most recent MODIS or VIIRS 1km L2P datum within 0.01 degrees from the grid point. \"Fill value\" indicates absence of such 1km data at the grid point.
Array Chunk Bytes 144.83 GiB 31.96 MiB Shape (30, 17999, 36000) (1, 1447, 2895) Count 10170 Tasks 5070 Chunks Type timedelta64[ns] numpy.ndarray - mask(time, lat, lon)float32dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
- long_name :
- sea/land field composite mask
- valid_min :
- 1
- valid_max :
- 31
- flag_masks :
- [1, 2, 4, 8, 16]
- flag_meanings :
- open_sea land open_lake open_sea_with_ice_in_the_grid open_lake_with_ice_in_the_grid
- comment :
- mask can be used to further filter the data.
- source :
- GMT \"grdlandmask\", ice flag from sea_ice_fraction data
Array Chunk Bytes 72.42 GiB 15.98 MiB Shape (30, 17999, 36000) (1, 1447, 2895) Count 10170 Tasks 5070 Chunks Type float32 numpy.ndarray - sea_ice_fraction(time, lat, lon)float32dask.array<chunksize=(1, 1447, 2895), meta=np.ndarray>
- long_name :
- sea ice area fraction
- standard_name :
- sea_ice_area_fraction
- valid_min :
- 0
- valid_max :
- 100
- source :
- EUMETSAT OSI-SAF, copyright EUMETSAT
- comment :
- ice fraction is a dimensionless quantity between 0 and 1; it has been interpolated by a nearest neighbor approach.
Array Chunk Bytes 72.42 GiB 15.98 MiB Shape (30, 17999, 36000) (1, 1447, 2895) Count 10170 Tasks 5070 Chunks Type float32 numpy.ndarray - sst_anomaly(time, lat, lon)float32dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
- long_name :
- SST anomaly from a seasonal SST climatology based on the MUR data over 2003-2014 period
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- anomaly reference to the day-of-year average between 2003 and 2014
Array Chunk Bytes 72.42 GiB 7.99 MiB Shape (30, 17999, 36000) (1, 1023, 2047) Count 19470 Tasks 9720 Chunks Type float32 numpy.ndarray
- Conventions :
- CF-1.7
- title :
- Daily MUR SST, Final product
- summary :
- A merged, multi-sensor L4 Foundation SST analysis product from JPL.
- references :
- http://podaac.jpl.nasa.gov/Multi-scale_Ultra-high_Resolution_MUR-SST
- institution :
- Jet Propulsion Laboratory
- history :
- created at nominal 4-day latency; replaced nrt (1-day latency) version.
- comment :
- MUR = \"Multi-scale Ultra-high Resolution\"
- license :
- These data are available free of charge under data policy of JPL PO.DAAC.
- id :
- MUR-JPL-L4-GLOB-v04.1
- naming_authority :
- org.ghrsst
- product_version :
- 04.1
- uuid :
- 27665bc0-d5fc-11e1-9b23-0800200c9a66
- gds_version_id :
- 2.0
- netcdf_version_id :
- 4.1
- date_created :
- 20210910T072132Z
- start_time :
- 20210901T090000Z
- stop_time :
- 20210901T090000Z
- time_coverage_start :
- 20210831T210000Z
- time_coverage_end :
- 20210901T210000Z
- file_quality_level :
- 3
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
- platform :
- Terra, Aqua, GCOM-W, MetOp-A, MetOp-B, Buoys/Ships
- sensor :
- MODIS, AMSR2, AVHRR, in-situ
- Metadata_Conventions :
- Unidata Observation Dataset v1.0
- metadata_link :
- http://podaac.jpl.nasa.gov/ws/metadata/dataset/?format=iso&shortName=MUR-JPL-L4-GLOB-v04.1
- keywords :
- Oceans > Ocean Temperature > Sea Surface Temperature
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Metadata Convention
- southernmost_latitude :
- -90.0
- northernmost_latitude :
- 90.0
- westernmost_longitude :
- -180.0
- easternmost_longitude :
- 180.0
- spatial_resolution :
- 0.01 degrees
- geospatial_lat_units :
- degrees north
- geospatial_lat_resolution :
- 0.009999999776
- geospatial_lon_units :
- degrees east
- geospatial_lon_resolution :
- 0.009999999776
- acknowledgment :
- Please acknowledge the use of these data with the following statement: These data were provided by JPL under support by NASA MEaSUREs program.
- creator_name :
- JPL MUR SST project
- creator_email :
- ghrsst@podaac.jpl.nasa.gov
- creator_url :
- http://mur.jpl.nasa.gov
- project :
- NASA Making Earth Science Data Records for Use in Research Environments (MEaSUREs) Program
- publisher_name :
- GHRSST Project Office
- publisher_url :
- http://www.ghrsst.org
- publisher_email :
- ghrsst-po@nceo.ac.uk
- processing_level :
- L4
- cdm_data_type :
- grid
Look at the Analysed SST variable metadata
all_sst = ds.analysed_sst
all_sst<xarray.DataArray 'analysed_sst' (time: 30, lat: 17999, lon: 36000)>
dask.array<concatenate, shape=(30, 17999, 36000), dtype=float32, chunksize=(1, 1023, 2047), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 -89.99 -89.98 -89.97 -89.96 ... 89.97 89.98 89.99
* lon (lon) float32 -180.0 -180.0 -180.0 -180.0 ... 180.0 180.0 180.0
* time (time) datetime64[ns] 2021-09-01T09:00:00 ... 2021-09-30T09:00:00
Attributes:
long_name: analysed sea surface temperature
standard_name: sea_surface_foundation_temperature
units: kelvin
valid_min: -32767
valid_max: 32767
comment: \"Final\" version using Multi-Resolution Variational Anal...
source: MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, A...- time: 30
- lat: 17999
- lon: 36000
- dask.array<chunksize=(1, 1023, 2047), meta=np.ndarray>
Array Chunk Bytes 72.42 GiB 7.99 MiB Shape (30, 17999, 36000) (1, 1023, 2047) Count 19470 Tasks 9720 Chunks Type float32 numpy.ndarray - lat(lat)float32-89.99 -89.98 ... 89.98 89.99
- long_name :
- latitude
- standard_name :
- latitude
- axis :
- Y
- units :
- degrees_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- comment :
- geolocations inherited from the input data without correction
array([-89.99, -89.98, -89.97, ..., 89.97, 89.98, 89.99], dtype=float32)
- lon(lon)float32-180.0 -180.0 ... 180.0 180.0
- long_name :
- longitude
- standard_name :
- longitude
- axis :
- X
- units :
- degrees_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- comment :
- geolocations inherited from the input data without correction
array([-179.99, -179.98, -179.97, ..., 179.98, 179.99, 180. ], dtype=float32) - time(time)datetime64[ns]2021-09-01T09:00:00 ... 2021-09-...
- long_name :
- reference time of sst field
- standard_name :
- time
- axis :
- T
- comment :
- Nominal time of analyzed fields
array(['2021-09-01T09:00:00.000000000', '2021-09-02T09:00:00.000000000', '2021-09-03T09:00:00.000000000', '2021-09-04T09:00:00.000000000', '2021-09-05T09:00:00.000000000', '2021-09-06T09:00:00.000000000', '2021-09-07T09:00:00.000000000', '2021-09-08T09:00:00.000000000', '2021-09-09T09:00:00.000000000', '2021-09-10T09:00:00.000000000', '2021-09-11T09:00:00.000000000', '2021-09-12T09:00:00.000000000', '2021-09-13T09:00:00.000000000', '2021-09-14T09:00:00.000000000', '2021-09-15T09:00:00.000000000', '2021-09-16T09:00:00.000000000', '2021-09-17T09:00:00.000000000', '2021-09-18T09:00:00.000000000', '2021-09-19T09:00:00.000000000', '2021-09-20T09:00:00.000000000', '2021-09-21T09:00:00.000000000', '2021-09-22T09:00:00.000000000', '2021-09-23T09:00:00.000000000', '2021-09-24T09:00:00.000000000', '2021-09-25T09:00:00.000000000', '2021-09-26T09:00:00.000000000', '2021-09-27T09:00:00.000000000', '2021-09-28T09:00:00.000000000', '2021-09-29T09:00:00.000000000', '2021-09-30T09:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- analysed sea surface temperature
- standard_name :
- sea_surface_foundation_temperature
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- \"Final\" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
Create a dataset / variable that is only our area of interest and view its metadata
sst = ds.analysed_sst.sel(lat=lats, lon=lons)
sst<xarray.DataArray 'analysed_sst' (time: 30, lat: 801, lon: 1701)>
dask.array<getitem, shape=(30, 801, 1701), dtype=float32, chunksize=(1, 601, 1536), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float32 41.0 41.01 41.02 41.03 ... 48.97 48.98 48.99 49.0
* lon (lon) float32 -93.0 -92.99 -92.98 -92.97 ... -76.02 -76.01 -76.0
* time (time) datetime64[ns] 2021-09-01T09:00:00 ... 2021-09-30T09:00:00
Attributes:
long_name: analysed sea surface temperature
standard_name: sea_surface_foundation_temperature
units: kelvin
valid_min: -32767
valid_max: 32767
comment: \"Final\" version using Multi-Resolution Variational Anal...
source: MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, A...- time: 30
- lat: 801
- lon: 1701
- dask.array<chunksize=(1, 200, 1536), meta=np.ndarray>
Array Chunk Bytes 155.93 MiB 3.52 MiB Shape (30, 801, 1701) (1, 601, 1536) Count 19590 Tasks 120 Chunks Type float32 numpy.ndarray - lat(lat)float3241.0 41.01 41.02 ... 48.99 49.0
- long_name :
- latitude
- standard_name :
- latitude
- axis :
- Y
- units :
- degrees_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- comment :
- geolocations inherited from the input data without correction
array([41. , 41.01, 41.02, ..., 48.98, 48.99, 49. ], dtype=float32)
- lon(lon)float32-93.0 -92.99 ... -76.01 -76.0
- long_name :
- longitude
- standard_name :
- longitude
- axis :
- X
- units :
- degrees_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- comment :
- geolocations inherited from the input data without correction
array([-93. , -92.99, -92.98, ..., -76.02, -76.01, -76. ], dtype=float32)
- time(time)datetime64[ns]2021-09-01T09:00:00 ... 2021-09-...
- long_name :
- reference time of sst field
- standard_name :
- time
- axis :
- T
- comment :
- Nominal time of analyzed fields
array(['2021-09-01T09:00:00.000000000', '2021-09-02T09:00:00.000000000', '2021-09-03T09:00:00.000000000', '2021-09-04T09:00:00.000000000', '2021-09-05T09:00:00.000000000', '2021-09-06T09:00:00.000000000', '2021-09-07T09:00:00.000000000', '2021-09-08T09:00:00.000000000', '2021-09-09T09:00:00.000000000', '2021-09-10T09:00:00.000000000', '2021-09-11T09:00:00.000000000', '2021-09-12T09:00:00.000000000', '2021-09-13T09:00:00.000000000', '2021-09-14T09:00:00.000000000', '2021-09-15T09:00:00.000000000', '2021-09-16T09:00:00.000000000', '2021-09-17T09:00:00.000000000', '2021-09-18T09:00:00.000000000', '2021-09-19T09:00:00.000000000', '2021-09-20T09:00:00.000000000', '2021-09-21T09:00:00.000000000', '2021-09-22T09:00:00.000000000', '2021-09-23T09:00:00.000000000', '2021-09-24T09:00:00.000000000', '2021-09-25T09:00:00.000000000', '2021-09-26T09:00:00.000000000', '2021-09-27T09:00:00.000000000', '2021-09-28T09:00:00.000000000', '2021-09-29T09:00:00.000000000', '2021-09-30T09:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- analysed sea surface temperature
- standard_name :
- sea_surface_foundation_temperature
- units :
- kelvin
- valid_min :
- -32767
- valid_max :
- 32767
- comment :
- \"Final\" version using Multi-Resolution Variational Analysis (MRVA) method for interpolation
- source :
- MODIS_T-JPL, MODIS_A-JPL, AMSR2-REMSS, AVHRRMTA_G-NAVO, AVHRRMTB_G-NAVO, iQUAM-NOAA/NESDIS, Ice_Conc-OSISAF
XArray reads data lazily, i.e. only when our code actually needs it. Up to this point, we haven’t read any data values, only metadata. The next line will force XArray to read the portions of the source files containing our area of interest. Behind the scenes, the eosdis-zarr-store library is ensuring data is fetched as efficiently as possible.
Note: This line isn’t strictly necessary, since XArray will automatically read the data we need the first time our code tries to use it, but calling this will make sure that we can read the data multiple times later on without re-fetching anything from the source files.
This line will take several seconds to complete, but since it is retrieving only about 50 MB of data from 22 GB of source files, several seconds constitutes a significant time, bandwidth, and disk space savings.
sst.load();Now we can start looking at aggregations across the time dimension. In this case, plot the standard deviation of the temperature at each point to get a visual sense of how much temperatures fluctuate over the course of the month.
# We expect a warning here, from finding the standard deviation of arrays that contain all N/A values.
# numpy produces N/A for these points, though, which is exactly what we want.
stdev_sst = sst.std('time')
stdev_sst.name = 'stdev of analysed_sst [Kelvin]'
stdev_sst.plot();/srv/conda/envs/notebook/lib/python3.9/site-packages/numpy/lib/nanfunctions.py:1670: RuntimeWarning: Degrees of freedom <= 0 for slice.
var = nanvar(a, axis=axis, dtype=dtype, out=out, ddof=ddof,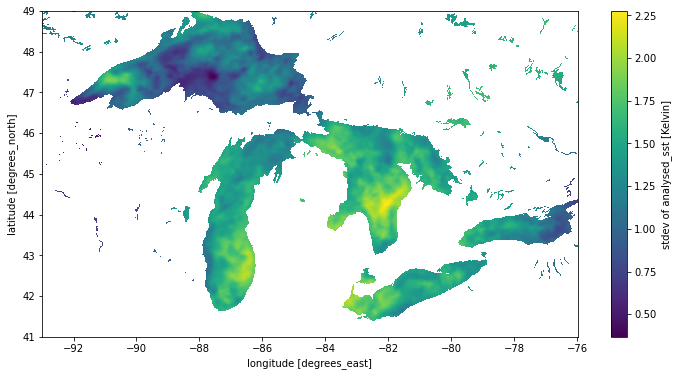
Interactive animation of a month of data
This section isn’t as important to fully understand. It shows us a way to get an interactive animation to see what we have retrieved so far
Define an animation function to plot the ith time step. We need to make sure each plot is using the same color scale, set by vmin and vmax so the animation is consistent
sst_min = sst.min()
sst_max = sst.max()
def show_time_step(i):
plt.clf()
res = sst[i].plot.imshow(vmin=sst_min, vmax=sst_max)
return (res,)Render each time slice once and show it as an HTML animation with interactive controls
#anim = animation.FuncAnimation(plt.gcf(), func=show_time_step, frames=len(sst))
#display(HTML(anim.to_jshtml()))
#plt.close()Supplemental: What’s happening here?
For EOSDIS data in the cloud, we have begun producing a metadata sidecar file in a format called DMR++ that extracts all of the information about arrays, variables, and dimensions from data files, as well as the byte offsets in the NetCDF4 file where data can be found. This information is sufficient to let the Zarr library read data from our NetCDF4 files, but it’s in the wrong format. zarr-eosdis-store knows how to fetch the sidecar file and transform it into something the Zarr library understands. Passing it when reading Zarr using XArray or the Zarr library lets these libraries interact with EOSDIS data exactly as if they were Zarr stores in a way that’s more optimal for reading data in the cloud. Beyond this, the zarr-eosdis-store library makes some optimizations in the way it reads data to help make up for situations where the NetCDF4 file is not internally arranged well for cloud-based access patterns.
