import glob
import numpy as np
import pandas as pd
import xarray as xr
import hvplot.xarray
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy
from datetime import datetime
import os
from os.path import isfile, basename, abspath
import dask
dask.config.set({"array.slicing.split_large_chunks": False})
import earthaccess
from earthaccess import Auth, DataCollections, DataGranules, StoreFrom the PO.DAAC Cookbook, to access the GitHub version of the notebook, follow this link.
Amazon Estuary Exploration:
Cloud Direct Download Version
This tutorial is one of two jupyter notebook versions of the same use case exploring multiple satellite data products over the Amazon Estuary. In this version, we use data that has been downloaded onto our local machine from the cloud.
Learning Objectives
- Compare cloud access methods (in tandem with notebook “Amazon Estuary Exploration: In Cloud AWS Version”)
- Search for data products using
earthaccessPython library - Access datasets using xarray and visualize using hvplot or plot tools
This tutorial explores the relationships between river height, land water equivalent thickness, sea surface salinity, and sea surface temperature in the Amazon River estuary and coastal region from multiple datasets listed below. The contents are useful for the ocean, coastal, and terrestrial hydrosphere communities, showcasing how to use cloud datasets and services. This notebook is meant to be executed locally.
Cloud Datasets
The tutorial itself will use four different datasets:
1. TELLUS_GRAC-GRFO_MASCON_CRI_GRID_RL06.1_V3
The Gravity Recovery And Climate Experiment Follow-On (GRACE-FO) satellite land water equivalent (LWE) thicknesses will be used to observe seasonal changes in water storage around the river. When discharge is high, the change in water storage will increase, thus highlighting a wet season. 2. PRESWOT_HYDRO_GRRATS_L2_DAILY_VIRTUAL_STATION_HEIGHTS_V2
The NASA Pre-SWOT Making Earth System Data Records for Use in Research Environments (MEaSUREs) Program virtual gauges will be used as a proxy for Surface Water and Ocean Topography (SWOT) discharge until SWOT products are available. MEaSUREs contains river height products, not discharge, but river height is directly related to discharge and thus will act as a good substitute.3. OISSS_L4_multimission_7day_v1
Optimally Interpolated Sea surface salinity (OISSS) is a level 4 product that combines the records from Aquarius (Sept 2011-June 2015), the Soil Moisture Active Passive (SMAP) satellite (April 2015-present), and ESAs Soil Moisture Ocean Salinity (SMOS) data to fill in data gaps.4. MODIS_AQUA_L3_SST_MID-IR_MONTHLY_9KM_NIGHTTIME_V2019.0
Sea surface temperature is obtained from the Moderate Resolution Imaging Spectrometer (MODIS) instrument on board the Aqua satellite. More details on available collections are on the PO.DAAC Cloud Earthdata Search Portal. For more information on the PO.DAAC transition to the cloud, please visit: https://podaac.jpl.nasa.gov/cloud-datasets/about
Note: NASA Earthdata Login Required
An Earthdata Login account is required to access data, as well as discover restricted data, from the NASA Earthdata system. Thus, to access NASA data, you need Earthdata Login. Please visit https://urs.earthdata.nasa.gov to register and manage your Earthdata Login account. This account is free to create and only takes a moment to set up. We use earthaccess to authenticate your login credentials below.
Needed Packages
auth = earthaccess.login(strategy="interactive", persist=True)Liquid Water Equivalent (LWE) Thickness (GRACE & GRACE-FO)
Search for GRACE LWE Thickness data
Suppose we are interested in LWE data from the dataset (DOI:10.5067/TEMSC-3JC62) described on this PO.DAAC dataset landing page: https://podaac.jpl.nasa.gov/dataset/TELLUS_GRAC-GRFO_MASCON_CRI_GRID_RL06.1_V3
From the landing page, we see the dataset Short Name under the Information tab. (For this dataset it is “TELLUS_GRAC-GRFO_MASCON_CRI_GRID_RL06.1_V3”) We will be using this to search for the necessary granules.
#earthaccess search
grace_results = earthaccess.search_data(short_name="TELLUS_GRAC-GRFO_MASCON_CRI_GRID_RL06.1_V3")Granules found: 1#download data into folder on local machine
earthaccess.download(grace_results, "./grace_data") Getting 1 granules, approx download size: 0.0 GB['GRCTellus.JPL.200204_202303.GLO.RL06.1M.MSCNv03CRI.nc']Open file using xarray.
#open dataset for visualization
ds_GRACE = xr.open_mfdataset("./grace_data/GRCTellus.JPL.200204_202303.GLO.RL06.1M.MSCNv03CRI.nc", engine="h5netcdf")
ds_GRACE<xarray.Dataset>
Dimensions: (lon: 720, lat: 360, time: 219, bounds: 2)
Coordinates:
* lon (lon) float64 0.25 0.75 1.25 1.75 ... 358.2 358.8 359.2 359.8
* lat (lat) float64 -89.75 -89.25 -88.75 ... 88.75 89.25 89.75
* time (time) datetime64[ns] 2002-04-17T12:00:00 ... 2023-03-16T1...
Dimensions without coordinates: bounds
Data variables:
lwe_thickness (time, lat, lon) float64 dask.array<chunksize=(219, 360, 720), meta=np.ndarray>
uncertainty (time, lat, lon) float64 dask.array<chunksize=(219, 360, 720), meta=np.ndarray>
lat_bounds (lat, bounds) float64 dask.array<chunksize=(360, 2), meta=np.ndarray>
lon_bounds (lon, bounds) float64 dask.array<chunksize=(720, 2), meta=np.ndarray>
time_bounds (time, bounds) datetime64[ns] dask.array<chunksize=(219, 2), meta=np.ndarray>
land_mask (lat, lon) float64 dask.array<chunksize=(360, 720), meta=np.ndarray>
scale_factor (lat, lon) float64 dask.array<chunksize=(360, 720), meta=np.ndarray>
mascon_ID (lat, lon) float64 dask.array<chunksize=(360, 720), meta=np.ndarray>
Attributes: (12/53)
Conventions: CF-1.6, ACDD-1.3, ISO 8601
Metadata_Conventions: Unidata Dataset Discovery v1.0
standard_name_vocabulary: NetCDF Climate and Forecast (CF) Metadata ...
title: JPL GRACE and GRACE-FO MASCON RL06.1Mv03 CRI
summary: Monthly gravity solutions from GRACE and G...
keywords: Solid Earth, Geodetics/Gravity, Gravity, l...
... ...
C_30_substitution: TN-14; Loomis et al., 2019, Geophys. Res. ...
user_note_1: The accelerometer on the GRACE-B spacecraf...
user_note_2: The accelerometer on the GRACE-D spacecraf...
journal_reference: Watkins, M. M., D. N. Wiese, D.-N. Yuan, C...
CRI_filter_journal_reference: Wiese, D. N., F. W. Landerer, and M. M. Wa...
date_created: 2023-05-22T06:05:03ZPlot a subset of the data
Use the function xarray.DataSet.sel to select a subset of the data to plot with hvplot.
lat_bnds, lon_bnds = [-18, 10], [275, 330] #degrees east for longitude
ds_GRACE_subset = ds_GRACE.sel(lat=slice(*lat_bnds), lon=slice(*lon_bnds))
ds_GRACE_subset
ds_GRACE_subset.lwe_thickness.hvplot.image(y='lat', x='lon', cmap='bwr_r',).opts(clim=(-80,80))River heights (Pre-SWOT MEaSUREs)
The shortname for MEaSUREs is ‘PRESWOT_HYDRO_GRRATS_L2_DAILY_VIRTUAL_STATION_HEIGHTS_V2’.
Our desired variable is height (meters above EGM2008 geoid) for this exercise, which can be subset by distance and time. Distance represents the distance from the river mouth, in this example, the Amazon estuary. Time is between April 8, 1993 and April 20, 2019.
To get the data for the exact area we need, we have set the boundaries of (-74.67188,-4.51279,-51.04688,0.19622) as reflected in our earthaccess data search.
MEaSUREs_results = earthaccess.search_data(short_name="PRESWOT_HYDRO_GRRATS_L2_DAILY_VIRTUAL_STATION_HEIGHTS_V2", temporal = ("1993-04-08", "2019-04-20"), bounding_box=(-74.67188,-4.51279,-51.04688,0.19622))Granules found: 1earthaccess.download(MEaSUREs_results, "./MEaSUREs_data") Getting 1 granules, approx download size: 0.0 GB['South_America_Amazon1kmdaily.nc']ds_MEaSUREs = xr.open_dataset("./MEaSUREs_data/South_America_Amazon1kmdaily.nc", engine="h5netcdf")
ds_MEaSUREs<xarray.Dataset>
Dimensions: (X: 3311, Y: 3311, distance: 3311, time: 9469,
charlength: 26)
Coordinates:
* time (time) datetime64[ns] 1993-04-08T15:20:40.665117184 ...
Dimensions without coordinates: X, Y, distance, charlength
Data variables:
lon (X) float64 ...
lat (Y) float64 ...
FD (distance) float64 ...
height (distance, time) float64 ...
sat (charlength, time) |S1 ...
storage (distance, time) float64 ...
IceFlag (time) float64 ...
LakeFlag (distance) float64 ...
Storage_uncertainty (distance, time) float64 ...
Attributes: (12/40)
title: GRRATS (Global River Radar Altimetry Time ...
Conventions: CF-1.6, ACDD-1.3
institution: Ohio State University, School of Earth Sci...
source: MEaSUREs OSU Storage toolbox 2018
keywords: EARTH SCIENCE,TERRESTRIAL HYDROSPHERE,SURF...
keywords_vocabulary: Global Change Master Directory (GCMD)
... ...
geospatial_lat_max: -0.6550700975069503
geospatial_lat_units: degree_north
geospatial_vertical_max: 92.7681246287056
geospatial_vertical_min: -3.5634095181633763
geospatial_vertical_units: m
geospatial_vertical_positive: upPlot a subset of the data
Plotting the river distances and associated heights on the map at time t=9069 (March 16, 2018) using plt.
fig = plt.figure(figsize=[11,7])
ax = plt.axes(projection=ccrs.PlateCarree())
ax.coastlines()
ax.set_extent([-85, -30, -20, 20])
ax.add_feature(cartopy.feature.RIVERS)
plt.scatter(ds_MEaSUREs.lon, ds_MEaSUREs.lat, lw=1, c=ds_MEaSUREs.height[:,9069])
plt.colorbar(label='Interpolated River Heights (m)')
plt.clim(-10,100)
plt.show()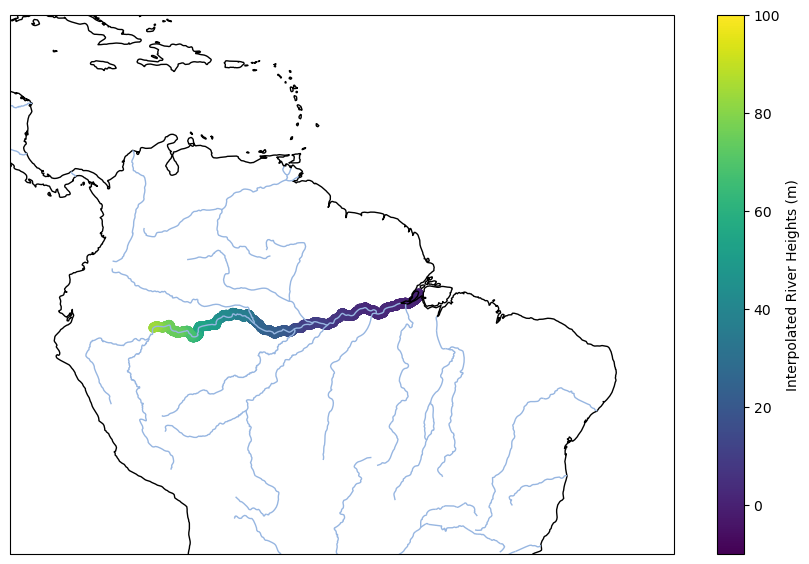
Sea Surface Salinity (Multi-mission: SMAP, Aquarius, SMOS)
The shortname for this dataset is ‘OISSS_L4_multimission_7day_v1’. This dataset contains hundreds of granules, by using earthaccess search, we access 998 granules.
Since this dataset has more than 1 granule that we want to open for visualization, we have to establish the full file path in a different way. For the previous datasets, we could list the exact file, but that would be difficult to do with hundreds of granules. Therefore, the extra step to recurse through the directory to access all files.
#earthaccess search
sss_results = earthaccess.search_data(short_name="OISSS_L4_multimission_7day_v1")Granules found: 998#earthaccess download
sss_files = earthaccess.download(sss_results, "./sss_data") Getting 998 granules, approx download size: 0.0 GB#ensures that all files are included in the path
sss_path = [os.path.join("./sss_data", f)
for pth, dirs, files in os.walk("./sss_data") for f in files]ds_sss = xr.open_mfdataset(sss_path,
combine='by_coords',
mask_and_scale=True,
decode_cf=True,
chunks='auto',
engine='h5netcdf')
ds_sss<xarray.Dataset>
Dimensions: (longitude: 1440, latitude: 720, time: 998)
Coordinates:
* longitude (longitude) float32 -179.9 -179.6 ... 179.6 179.9
* latitude (latitude) float32 -89.88 -89.62 ... 89.62 89.88
* time (time) datetime64[ns] 2011-08-28 ... 2022-08-02
Data variables:
sss (latitude, longitude, time) float32 dask.array<chunksize=(720, 1440, 1), meta=np.ndarray>
sss_empirical_uncertainty (latitude, longitude, time) float32 dask.array<chunksize=(720, 1440, 962), meta=np.ndarray>
sss_uncertainty (latitude, longitude, time) float32 dask.array<chunksize=(720, 1440, 1), meta=np.ndarray>
Attributes: (12/42)
Conventions: CF-1.8, ACDD-1.3
standard_name_vocabulary: CF Standard Name Table v27
Title: Multi-Mission Optimally Interpolated Sea S...
Short_Name: OISSS_L4_multimission_7d_v1
Version: V1.0
Processing_Level: Level 4
... ...
geospatial_lat_resolution: 0.25
geospatial_lat_units: degrees_north
geospatial_lon_min: -180.0
geospatial_lon_max: 180.0
geospatial_lon_resolution: 0.25
geospatial_lon_units: degrees_eastPlot a subset of the data
Use the function xarray.DataSet.sel to select a subset of the data at the outlet of the Amazon to plot at time t=0 (August 28, 2011) with hvplot.
lat_bnds, lon_bnds = [-2, 6], [-52, -44]
ds_sss_subset = ds_sss.sel(latitude=slice(*lat_bnds), longitude=slice(*lon_bnds))
ds_sss_subset
ds_sss_subset.sss[:,:,0].hvplot() Sea Surface Temperature (MODIS)
MODIS has SST data with separate files on a monthly basis. By using earthaccess search with the shortname: “MODIS_AQUA_L3_SST_MID-IR_MONTHLY_9KM_NIGHTTIME_V2019.0”, and filtering by time to coincide when SSS started (2011), we access 143 granules.
Get a list of files so we can open them all at once, creating an xarray dataset using the open_mfdataset() function to “read in” all of the netCDF4 files in one call. MODIS does not have a built-in time variable like SSS, but it is subset by latitude and longitude coordinates. We need to combine the files using the nested format with a created ‘time’ dimension.
sss_results = earthaccess.search_data(short_name="MODIS_AQUA_L3_SST_MID-IR_MONTHLY_9KM_NIGHTTIME_V2019.0", temporal = ("2011-01-01", "2023-01-01"))Granules found: 143sss_files = earthaccess.download(sss_results, "./modis_data") Getting 143 granules, approx download size: 0.0 GBMODIS did not come with a time variable, so it needs to be extracted from the file names and added in the file preprocessing so files can be successfully concatenated.
#repeat this step since we are accessing multiple granules
modis_path = [os.path.join("./modis_data", f)
for pth, dirs, files in os.walk("./modis_data") for f in files]#function for time dimension added to each netCDF file
def preprocessing(ds):
file_name = ds.product_name
file_date = basename(file_name).split("_")[2][:6]
file_date_c = datetime.strptime(file_date, "%Y%m")
time_point = [file_date_c]
ds.coords['time'] = ('time', time_point) #expand the dimensions to include time
return ds
ds_MODIS = xr.open_mfdataset(modis_path, combine='by_coords', join='override', preprocess = preprocessing)
ds_MODIS<xarray.Dataset>
Dimensions: (time: 143, lat: 2160, lon: 4320, rgb: 3, eightbitcolor: 256)
Coordinates:
* lat (lat) float32 89.96 89.88 89.79 89.71 ... -89.79 -89.88 -89.96
* lon (lon) float32 -180.0 -179.9 -179.8 -179.7 ... 179.8 179.9 180.0
* time (time) datetime64[ns] 2011-01-01 2011-02-01 ... 2023-01-01
Dimensions without coordinates: rgb, eightbitcolor
Data variables:
sst4 (time, lat, lon) float32 dask.array<chunksize=(1, 2160, 4320), meta=np.ndarray>
qual_sst4 (time, lat, lon) float32 dask.array<chunksize=(1, 2160, 4320), meta=np.ndarray>
palette (time, lon, lat, rgb, eightbitcolor) uint8 dask.array<chunksize=(1, 4320, 2160, 3, 256), meta=np.ndarray>
Attributes: (12/59)
product_name: AQUA_MODIS.20110101_20110131.L3m.MO.SST...
instrument: MODIS
title: MODISA Level-3 Standard Mapped Image
project: Ocean Biology Processing Group (NASA/GS...
platform: Aqua
temporal_range: month
... ...
publisher_url: https://oceandata.sci.gsfc.nasa.gov
processing_level: L3 Mapped
cdm_data_type: grid
data_bins: 4834400
data_minimum: -1.635
data_maximum: 32.06999Plot a subset of the data
Use the function xarray.DataSet.sel to select a subset of the data at the outlet of the Amazon to plot with hvplot.
lat_bnds, lon_bnds = [6, -2], [-52, -44]
ds_MODIS_subset = ds_MODIS.sel(lat=slice(*lat_bnds), lon=slice(*lon_bnds))
ds_MODIS_subset.sst4.hvplot.image(y='lat', x='lon', cmap='viridis').opts(clim=(22,30))Time Series Comparison
Plot each dataset for the time period 2011-2019.
First, we need to average all pixels in the subset lat/lon per time for sea surface salinity and sea surface temperature to set up for the graphs.
sss_mean = []
for t in np.arange(len(ds_sss_subset.time)):
sss_mean.append(np.nanmean(ds_sss_subset.sss[:,:,t].values))
#sss_mean#MODIS
sst_MODIS_mean = []
for t in np.arange(len(ds_MODIS_subset.time)):
sst_MODIS_mean.append(np.nanmean(ds_MODIS_subset.sst4[t,:,:].values))
#sst_MODIS_meanCombined timeseries plot of river height and LWE thickness
Both datasets are mapped for the outlet of the Amazon River into the estuary.
#plot river height and land water equivalent thickness
fig, ax1 = plt.subplots(figsize=[12,7])
#plot river height
ds_MEaSUREs.height[16,6689:9469].plot(color='darkblue')
#plot LWE thickness on secondary axis
ax2 = ax1.twinx()
ax2.plot(ds_GRACE_subset.time[107:179], ds_GRACE_subset.lwe_thickness[107:179,34,69], color = 'darkorange')
ax1.set_xlabel('Date')
ax2.set_ylabel('Land Water Equivalent Thickness (cm)', color='darkorange')
ax1.set_ylabel('River Height (m)', color='darkblue')
ax2.legend(['GRACE-FO'], loc='upper right')
ax1.legend(['Pre-SWOT MEaSUREs'], loc='lower right')
plt.title('Amazon Estuary, 2011-2019 Lat, Lon = (-0.7, -50)')
ax1.grid()
plt.show()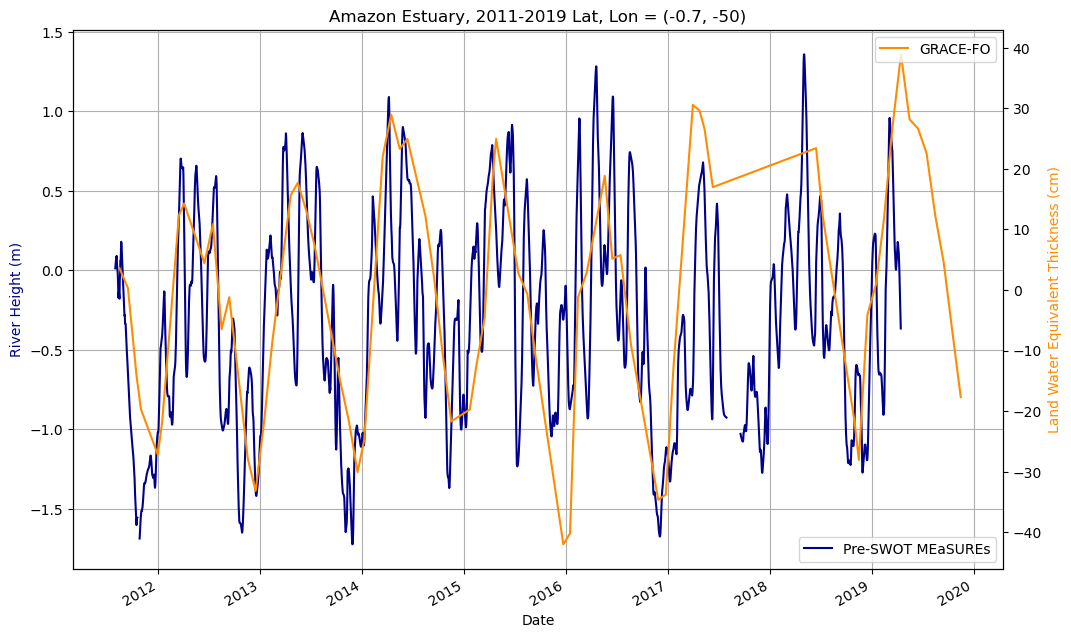
LWE thickness captures the seasonality of Pre-SWOT MEaSUREs river heights well, and so LWE thickness can be compared to all other variables as a representative of the seasonality of both measurements for the purpose of this notebook.
Combined timeseries plots of salinity and LWE thickness, followed by temperature
#Combined Subplots
fig = plt.figure(figsize=(10,10))
ax1 = fig.add_subplot(211)
plt.title('Amazon Estuary, 2011-2019')
ax2 = ax1.twinx()
ax3 = plt.subplot(212)
ax4 = ax3.twinx()
#lwe thickness
ax1.plot(ds_GRACE_subset.time[107:179], ds_GRACE_subset.lwe_thickness[107:179,34,69], color = 'darkorange')
ax1.set_ylabel('LWE Thickness (cm)', color='darkorange')
ax1.grid()
#sea surface salinity
ax2.plot(ds_sss_subset.time[0:750], sss_mean[0:750], 'g')
ax2.set_ylabel('SSS (psu)', color='g')
#sea surface temperature
ax3.plot(ds_MODIS_subset.time[7:108], sst_MODIS_mean[7:108], 'darkred')
ax3.set_ylabel('SST (deg C)', color='darkred')
ax3.grid()
#river height at outlet
ds_MEaSUREs.height[16,6689:9469].plot(color='darkblue')
ax4.set_ylabel('River Height (m)', color='darkblue')Text(0, 0.5, 'River Height (m)')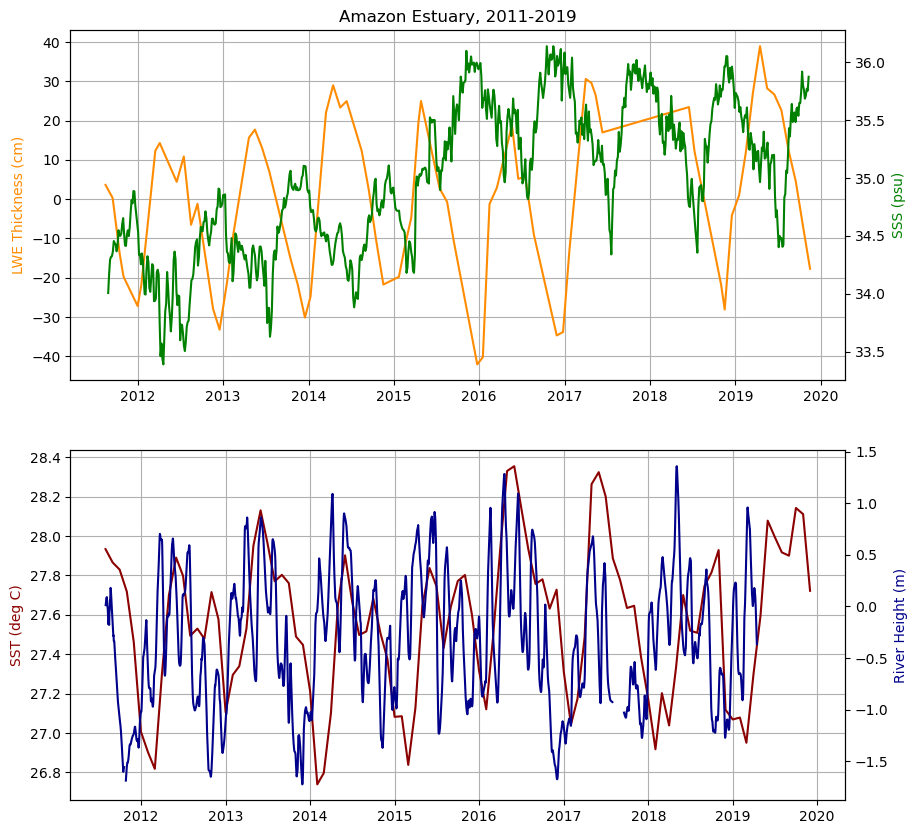
Measurements of LWE thickness and SSS follow expected patterns. When lwe thickness is at its lowest, indicating less water is flowing through during the drought, salinity is at its highest. Without high volume of water pouring into the estuary, salinity increases. We can see that temperature is shifted a bit in time from river height as well at the outlet, a relationship that could be further explored.

