import glob
import h5netcdf
import xarray as xr
import pandas as pd
import geopandas as gpd
import contextily as cx
import numpy as np
import matplotlib.pyplot as plt
import hvplot.xarray
import zipfile
import earthaccessFrom the PO.DAAC Cookbook, to access the GitHub version of the notebook, follow this link.
Note: This notebook uses Version C (2.0) of SWOT data that was available at the time of this notebook’s development. The most recent data is now available as Version D for SWOT collections. The last Version C measurement will be until May 3rd, 2025. The first Version D measurement starts on May 5th, 2025.
SWOT Hydrology Dataset Exploration on a local machine
Accessing and Visualizing SWOT Datasets
Requirement:
Local compute environment e.g. laptop, server: this tutorial can be run on your local machine.
Learning Objectives:
- Access SWOT HR data products (archived in NASA Earthdata Cloud) by downloading to local machine
- Visualize accessed data for a quick check
SWOT Level 2 KaRIn High Rate Version C (aka 2.0) Datasets:
River Vector Shapefile - SWOT_L2_HR_RIVERSP_2.0
Lake Vector Shapefile - SWOT_L2_HR_LAKESP_2.0
Water Mask Pixel Cloud NetCDF - SWOT_L2_HR_PIXC_2.0
Water Mask Pixel Cloud Vector Attribute NetCDF - SWOT_L2_HR_PIXCVec_2.0
Raster NetCDF - SWOT_L2_HR_Raster_2.0
Single Look Complex Data product - SWOT_L1B_HR_SLC_2.0
Notebook Author: Cassie Nickles, NASA PO.DAAC (Feb 2024) || Other Contributors: Zoe Walschots (PO.DAAC Summer Intern 2023), Catalina Taglialatela (NASA PO.DAAC), Luis Lopez (NASA NSIDC DAAC), Brent Williams (NASA JPL)
Last updated: 9 July 2024
Libraries Needed
Earthdata Login
An Earthdata Login account is required to access data, as well as discover restricted data, from the NASA Earthdata system. Thus, to access NASA data, you need Earthdata Login. If you don’t already have one, please visit https://urs.earthdata.nasa.gov to register and manage your Earthdata Login account. This account is free to create and only takes a moment to set up. We use earthaccess to authenticate your login credentials below.
auth = earthaccess.login()Single File Access
1. River Vector Shapefiles
The https access link can be found using earthaccess data search. Since this collection consists of Reach and Node files, we need to extract only the granule for the Reach file. We do this by filtering for the ‘Reach’ title in the data link.
Alternatively, Earthdata Search (see tutorial) can be used to manually search in a GUI interface.
For additional tips on spatial searching of SWOT HR L2 data, see also PO.DAAC Cookbook - SWOT Chapter tips section.
Search for the data of interest
#Retrieves granule from the day we want, in this case by passing to `earthdata.search_data` function the data collection shortname, temporal bounds, and filter by wildcards
river_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_RIVERSP_2.0',
temporal = ('2024-02-01 00:00:00', '2024-07-15 23:59:59'), # can also specify by time
granule_name = '*Reach*_287_NA*') # here we filter by Reach files (not node), pass=287, continent code=NAGranules found: 8Dowload, unzip, read the data
Let’s download the first data file! earthaccess.download has a list as the input format, so we need to put brackets around the single file we pass.
earthaccess.download([river_results[0]], "./data_downloads") Getting 1 granules, approx download size: 0.01 GB['data_downloads\\SWOT_L2_HR_RiverSP_Reach_010_287_NA_20240204T060400_20240204T060409_PIC0_01.zip']The native format for this data is a .zip file, and we want the .shp file within the .zip file, so we must first extract the data to open it. First, we’ll programmatically get the filename we just downloaded, and then extract all data to the data_downloads folder.
filename = earthaccess.results.DataGranule.data_links(river_results[0], access='external')
filename = filename[0].split("/")[-1]
filename'SWOT_L2_HR_RiverSP_Reach_010_287_NA_20240204T060400_20240204T060409_PIC0_01.zip'with zipfile.ZipFile(f'data_downloads/{filename}', 'r') as zip_ref:
zip_ref.extractall('data_downloads')Open the shapefile using geopandas
filename_shp = filename.replace('.zip','.shp')SWOT_HR_shp1 = gpd.read_file(f'data_downloads/{filename_shp}')
#view the attribute table
SWOT_HR_shp1 | reach_id | time | time_tai | time_str | p_lat | p_lon | river_name | wse | wse_u | wse_r_u | ... | p_wid_var | p_n_nodes | p_dist_out | p_length | p_maf | p_dam_id | p_n_ch_max | p_n_ch_mod | p_low_slp | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 71224800093 | -1.000000e+12 | -1.000000e+12 | no_data | 48.724265 | -92.406254 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 232341.227 | 90 | 47778.423 | 18013.132474 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-92.51093 48.70847, -92.51052 48.7... |
| 1 | 71224800101 | -1.000000e+12 | -1.000000e+12 | no_data | 48.739159 | -92.290054 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 767.700 | 6 | 48958.712 | 1180.288364 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-92.29723 48.73905, -92.29682 48.7... |
| 2 | 71224800114 | -1.000000e+12 | -1.000000e+12 | no_data | 48.743344 | -92.283320 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 2911.208 | 3 | 49549.648 | 590.936467 | -1.000000e+12 | 23000 | 2 | 1 | 0 | LINESTRING (-92.28569 48.74125, -92.28495 48.7... |
| 3 | 71224800123 | 7.603424e+08 | 7.603424e+08 | 2024-02-04T06:13:10Z | 48.751442 | -92.242669 | no_data | 3.585147e+02 | 2.006910e+00 | 2.004890e+00 | ... | 57688.777 | 31 | 55684.066 | 6134.417666 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-92.28196 48.74559, -92.28163 48.7... |
| 4 | 71224800133 | 7.603424e+08 | 7.603424e+08 | 2024-02-04T06:13:10Z | 48.762334 | -92.189341 | no_data | 3.579681e+02 | 1.451600e-01 | 1.138900e-01 | ... | 20821.463 | 13 | 58222.719 | 2538.653439 | -1.000000e+12 | 0 | 3 | 1 | 0 | LINESTRING (-92.20553 48.75837, -92.20512 48.7... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1058 | 77127000061 | 7.603418e+08 | 7.603419e+08 | 2024-02-04T06:04:09Z | 18.050684 | -98.761645 | no_data | 6.529558e+02 | 1.000896e+02 | 1.000896e+02 | ... | 784.041 | 67 | 667747.660 | 13493.202300 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-98.81280 18.06539, -98.81280 18.0... |
| 1059 | 77127000071 | -1.000000e+12 | -1.000000e+12 | no_data | 17.981704 | -98.686712 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 824.145 | 97 | 687123.984 | 19376.324005 | -1.000000e+12 | 0 | 3 | 1 | 0 | LINESTRING (-98.71239 18.03246, -98.71239 18.0... |
| 1060 | 77127000131 | 7.603418e+08 | 7.603419e+08 | 2024-02-04T06:04:09Z | 18.102586 | -98.771552 | no_data | 6.576003e+02 | 1.240586e+02 | 1.240586e+02 | ... | 281.012 | 77 | 683164.834 | 15417.173639 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-98.81280 18.06539, -98.81280 18.0... |
| 1061 | 77127000141 | -1.000000e+12 | -1.000000e+12 | no_data | 18.094132 | -98.694466 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 414.760 | 54 | 693896.634 | 10731.799933 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-98.71770 18.11625, -98.71764 18.1... |
| 1062 | 77127000151 | -1.000000e+12 | -1.000000e+12 | no_data | 18.097046 | -98.657280 | no_data | -1.000000e+12 | -1.000000e+12 | -1.000000e+12 | ... | 436.883 | 54 | 704624.208 | 10727.574606 | -1.000000e+12 | 0 | 2 | 1 | 0 | LINESTRING (-98.66628 18.07224, -98.66611 18.0... |
1063 rows × 127 columns
Quickly plot the SWOT river data
# Simple plot
fig, ax = plt.subplots(figsize=(7,5))
SWOT_HR_shp1.plot(ax=ax, color='black')
cx.add_basemap(ax, crs=SWOT_HR_shp1.crs, source=cx.providers.OpenTopoMap)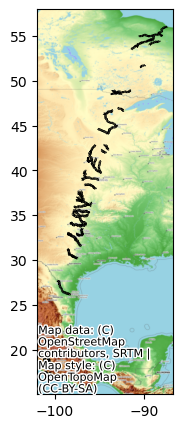
# Another way to plot geopandas dataframes is with `explore`, which also plots a basemap
#SWOT_HR_shp1.explore()2. Lake Vector Shapefiles
The lake vector shapefiles can be accessed in the same way as the river shapefiles above.
For additional tips on spatial searching of SWOT HR L2 data, see also PO.DAAC Cookbook - SWOT Chapter tips section.
Search for data of interest
lake_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_LAKESP_2.0',
temporal = ('2024-02-01 00:00:00', '2024-07-15 23:59:59'), # can also specify by time
granule_name = '*Prior*_287_NA*') # here we filter by files with 'Prior' in the name (This collection has three options: Obs, Unassigned, and Prior), pass 287 and continent code=NAGranules found: 8Let’s download the first data file! earthaccess.download has a list as the input format, so we need to put brackets around the single file we pass.
earthaccess.download([lake_results[0]], "./data_downloads") Getting 1 granules, approx download size: 0.07 GB['data_downloads\\SWOT_L2_HR_LakeSP_Prior_010_287_NA_20240204T060400_20240204T061541_PIC0_01.zip']The native format for this data is a .zip file, and we want the .shp file within the .zip file, so we must first extract the data to open it. First, we’ll programmatically get the filename we just downloaded, and then extract all data to the SWOT_downloads folder.
filename2 = earthaccess.results.DataGranule.data_links(lake_results[0], access='external')
filename2 = filename2[0].split("/")[-1]
filename2'SWOT_L2_HR_LakeSP_Prior_010_287_NA_20240204T060400_20240204T061541_PIC0_01.zip'with zipfile.ZipFile(f'data_downloads/{filename2}', 'r') as zip_ref:
zip_ref.extractall('data_downloads')Open the shapefile using geopandas
filename_shp2 = filename2.replace('.zip','.shp')
filename_shp2'SWOT_L2_HR_LakeSP_Prior_010_287_NA_20240204T060400_20240204T061541_PIC0_01.shp'SWOT_HR_shp2 = gpd.read_file(f'data_downloads/{filename_shp2}')
#view the attribute table
SWOT_HR_shp2| lake_id | reach_id | obs_id | overlap | n_overlap | time | time_tai | time_str | wse | wse_u | ... | lake_name | p_res_id | p_lon | p_lat | p_ref_wse | p_ref_area | p_date_t0 | p_ds_t0 | p_storage | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7120822822 | no_data | 712239R999998 | 99 | 1 | 7.603424e+08 | 7.603424e+08 | 2024-02-04T06:13:08Z | 5.281870e+02 | 1.500000e-02 | ... | no_data | -99999999 | -91.557528 | 47.616292 | -1.000000e+12 | 1.038600 | no_data | -1.000000e+12 | -1.000000e+12 | MULTIPOLYGON (((-91.56583 47.61200, -91.56589 ... |
| 1 | 7120822902 | no_data | no_data | no_data | no_data | -1.000000e+12 | -1.000000e+12 | no_data | -1.000000e+12 | -1.000000e+12 | ... | no_data | -99999999 | -91.623241 | 47.756499 | -1.000000e+12 | 0.113400 | no_data | -1.000000e+12 | -1.000000e+12 | None |
| 2 | 7120822932 | no_data | no_data | no_data | no_data | -1.000000e+12 | -1.000000e+12 | no_data | -1.000000e+12 | -1.000000e+12 | ... | TONY LAKE | -99999999 | -91.635242 | 47.726123 | -1.000000e+12 | 0.017100 | no_data | -1.000000e+12 | -1.000000e+12 | None |
| 3 | 7120822982 | no_data | no_data | no_data | no_data | -1.000000e+12 | -1.000000e+12 | no_data | -1.000000e+12 | -1.000000e+12 | ... | no_data | -99999999 | -91.665522 | 47.705366 | -1.000000e+12 | 0.026100 | no_data | -1.000000e+12 | -1.000000e+12 | None |
| 4 | 7120823182 | no_data | no_data | no_data | no_data | -1.000000e+12 | -1.000000e+12 | no_data | -1.000000e+12 | -1.000000e+12 | ... | HEART LAKE | -99999999 | -91.651807 | 47.769148 | -1.000000e+12 | 0.124200 | no_data | -1.000000e+12 | -1.000000e+12 | None |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 65495 | 7130133552 | no_data | 713244L999972;713244L000002 | 73;2 | 2 | 7.603424e+08 | 7.603424e+08 | 2024-02-04T06:14:01Z | 3.926960e+02 | 5.900000e-02 | ... | no_data | -99999999 | -90.889026 | 50.669027 | -1.000000e+12 | 0.695690 | no_data | -1.000000e+12 | -1.000000e+12 | POLYGON ((-90.89210 50.67709, -90.89185 50.677... |
| 65496 | 7130141612 | no_data | 713245L999974;713245L000001 | 31;5 | 2 | 7.603424e+08 | 7.603425e+08 | 2024-02-04T06:14:08Z | 3.741850e+02 | 8.200000e-02 | ... | LAKE ST JOSEPH;ST JOSEPH | -99999999 | -90.750682 | 51.061383 | -1.000000e+12 | 0.256500 | no_data | -1.000000e+12 | -1.000000e+12 | MULTIPOLYGON (((-90.75251 51.06305, -90.75228 ... |
| 65497 | 7420206383 | 74226000013;74227100043;74227100013;7422710006... | 742214L000175;742214L999934;742214L000500;7422... | 64;23;0;0 | 4 | 7.603421e+08 | 7.603422e+08 | 2024-02-04T06:09:01Z | 1.875740e+02 | 1.000000e-03 | ... | LAKE TEXOMA | 1135 | -96.688976 | 33.901142 | -1.000000e+12 | 257.028517 | no_data | -1.000000e+12 | -1.000000e+12 | MULTIPOLYGON (((-96.70899 33.82534, -96.70885 ... |
| 65498 | 7420280413 | 74246000423;74246000413;74246000404 | 742218L999996;742218L001654;742219L999885 | 4;0;0 | 3 | 7.603422e+08 | 7.603422e+08 | 2024-02-04T06:09:48Z | 1.941800e+02 | 2.100000e-02 | ... | OOLAGAHL LAKE;OOLOGAH LAKE | 1032 | -95.593848 | 36.550604 | -1.000000e+12 | 123.796498 | no_data | -1.000000e+12 | -1.000000e+12 | MULTIPOLYGON (((-95.67217 36.43803, -95.67157 ... |
| 65499 | 7710056183 | 77125000273;77125000263;77125000283;7712500030... | 771186L999995;771186L000013;771186L999993 | 12;1;0 | 3 | 7.603419e+08 | 7.603419e+08 | 2024-02-04T06:04:22Z | 4.586500e+02 | 3.400000e-02 | ... | PRESA EL CARACOL | 1384 | -99.861530 | 17.975030 | -1.000000e+12 | 35.410155 | no_data | -1.000000e+12 | -1.000000e+12 | MULTIPOLYGON (((-99.75753 18.02115, -99.75739 ... |
65500 rows × 51 columns
Quickly plot the SWOT lakes data
fig, ax = plt.subplots(figsize=(7,5))
SWOT_HR_shp2.plot(ax=ax, color='black')
cx.add_basemap(ax, crs=SWOT_HR_shp2.crs, source=cx.providers.OpenTopoMap)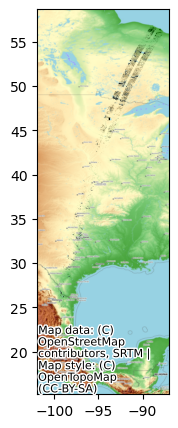
Accessing the remaining files is different than the shp files above. We do not need to extract the shapefiles from a zip file because the following SWOT HR collections are stored in netCDF files in the cloud. For the rest of the products, we will open via xarray, not geopandas.
3. Water Mask Pixel Cloud NetCDF
Search for data collection and time of interest
pixc_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_PIXC_2.0',
temporal = ('2024-01-25 00:00:00', '2024-05-29 23:59:59'), # can also specify by time
bounding_box = (-106.62, 38.809, -106.54, 38.859)) # Lake Travis near Austin, TXGranules found: 27Let’s download one data file! earthaccess.download has a list as the input format, so we need to put brackets around the single file we pass.
earthaccess.download([pixc_results[0]], "./data_downloads") Getting 1 granules, approx download size: 0.37 GB['data_downloads\\SWOT_L2_HR_PIXC_010_106_086R_20240128T183540_20240128T183551_PIC0_01.nc']Open data using xarray
The pixel cloud netCDF files are formatted with three groups titled, “pixel cloud”, “tvp”, or “noise” (more detail here). In order to access the coordinates and variables within the file, a group must be specified when calling xarray open_dataset.
ds_PIXC = xr.open_mfdataset("data_downloads/SWOT_L2_HR_PIXC_*.nc", group = 'pixel_cloud', engine='h5netcdf')
ds_PIXC<xarray.Dataset>
Dimensions: (points: 3959179, complex_depth: 2,
num_pixc_lines: 3278)
Coordinates:
latitude (points) float64 dask.array<chunksize=(3959179,), meta=np.ndarray>
longitude (points) float64 dask.array<chunksize=(3959179,), meta=np.ndarray>
Dimensions without coordinates: points, complex_depth, num_pixc_lines
Data variables: (12/61)
azimuth_index (points) float64 dask.array<chunksize=(3959179,), meta=np.ndarray>
range_index (points) float64 dask.array<chunksize=(3959179,), meta=np.ndarray>
interferogram (points, complex_depth) float32 dask.array<chunksize=(3959179, 2), meta=np.ndarray>
power_plus_y (points) float32 dask.array<chunksize=(3959179,), meta=np.ndarray>
power_minus_y (points) float32 dask.array<chunksize=(3959179,), meta=np.ndarray>
coherent_power (points) float32 dask.array<chunksize=(3959179,), meta=np.ndarray>
... ...
pixc_line_qual (num_pixc_lines) float64 dask.array<chunksize=(3278,), meta=np.ndarray>
pixc_line_to_tvp (num_pixc_lines) float32 dask.array<chunksize=(3278,), meta=np.ndarray>
data_window_first_valid (num_pixc_lines) float64 dask.array<chunksize=(3278,), meta=np.ndarray>
data_window_last_valid (num_pixc_lines) float64 dask.array<chunksize=(3278,), meta=np.ndarray>
data_window_first_cross_track (num_pixc_lines) float32 dask.array<chunksize=(3278,), meta=np.ndarray>
data_window_last_cross_track (num_pixc_lines) float32 dask.array<chunksize=(3278,), meta=np.ndarray>
Attributes:
description: cloud of geolocated interferogram pixels
interferogram_size_azimuth: 3278
interferogram_size_range: 5718
looks_to_efflooks: 1.550665505987333
num_azimuth_looks: 7.0
azimuth_offset: 6For plotting PIXC using classification and quality flags
# mask to get good water pixels
mask = np.where(np.logical_and(ds_PIXC.classification > 2, ds_PIXC.geolocation_qual <28800))
#For more conservative filtering, use ds_PIXC.geolocation_qual <4 to also remove suspect values
plt.scatter(x=ds_PIXC.longitude[mask], y=ds_PIXC.latitude[mask], c=ds_PIXC.height[mask])
plt.colorbar().set_label('Height (m)')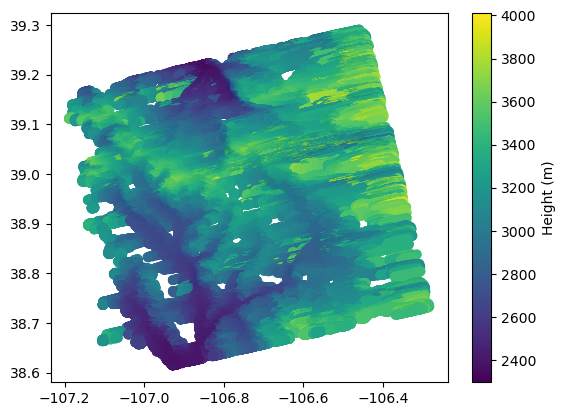
4. Water Mask Pixel Cloud Vector Attribute NetCDF
Search for data of interest
#Let's plot the same pass and tile as the above
pixcvec_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_PIXCVEC_2.0',
granule_name = '*010_106_086R*') #The same cycle, pass and tile as previously downloadedGranules found: 1Let’s download the first data file! earthaccess.download has a list as the input format, so we need to put brackets around the single file we pass.
earthaccess.download([pixcvec_results[0]], "./data_downloads") Getting 1 granules, approx download size: 0.29 GB['data_downloads\\SWOT_L2_HR_PIXCVec_010_106_086R_20240128T183540_20240128T183551_PIC0_01.nc']Open data using xarray
First, we’ll programmatically get the filename we just downloaded and then view the file via xarray.
ds_PIXCVEC = xr.open_mfdataset("data_downloads/SWOT_L2_HR_PIXCVec_*.nc", decode_cf=False, engine='h5netcdf')
ds_PIXCVEC<xarray.Dataset>
Dimensions: (points: 3959179, nchar_reach_id: 11,
nchar_node_id: 14, nchar_lake_id: 10,
nchar_obs_id: 13)
Dimensions without coordinates: points, nchar_reach_id, nchar_node_id,
nchar_lake_id, nchar_obs_id
Data variables:
azimuth_index (points) int32 dask.array<chunksize=(3959179,), meta=np.ndarray>
range_index (points) int32 dask.array<chunksize=(3959179,), meta=np.ndarray>
latitude_vectorproc (points) float64 dask.array<chunksize=(3959179,), meta=np.ndarray>
longitude_vectorproc (points) float64 dask.array<chunksize=(3959179,), meta=np.ndarray>
height_vectorproc (points) float32 dask.array<chunksize=(3959179,), meta=np.ndarray>
reach_id (points, nchar_reach_id) |S1 dask.array<chunksize=(3959179, 11), meta=np.ndarray>
node_id (points, nchar_node_id) |S1 dask.array<chunksize=(3959179, 14), meta=np.ndarray>
lake_id (points, nchar_lake_id) |S1 dask.array<chunksize=(3959179, 10), meta=np.ndarray>
obs_id (points, nchar_obs_id) |S1 dask.array<chunksize=(3959179, 13), meta=np.ndarray>
ice_clim_f (points) int8 dask.array<chunksize=(3959179,), meta=np.ndarray>
ice_dyn_f (points) int8 dask.array<chunksize=(3959179,), meta=np.ndarray>
Attributes: (12/45)
Conventions: CF-1.7
title: Level 2 KaRIn high rate pixel cloud vect...
short_name: L2_HR_PIXCVec
institution: CNES
source: Level 1B KaRIn High Rate Single Look Com...
history: 2024-02-02T11:20:25.557946Z: Creation
... ...
xref_prior_river_db_file:
xref_prior_lake_db_file: SWOT_LakeDatabase_Nom_106_20000101T00000...
xref_reforbittrack_files: SWOT_RefOrbitTrackTileBoundary_Nom_20000...
xref_param_l2_hr_laketile_file: SWOT_Param_L2_HR_LakeTile_20000101T00000...
ellipsoid_semi_major_axis: 6378137.0
ellipsoid_flattening: 0.0033528106647474805Simple plot
pixcvec_htvals = ds_PIXCVEC.height_vectorproc.compute()
pixcvec_latvals = ds_PIXCVEC.latitude_vectorproc.compute()
pixcvec_lonvals = ds_PIXCVEC.longitude_vectorproc.compute()
#Before plotting, we set all fill values to nan so that the graph shows up better spatially
pixcvec_htvals[pixcvec_htvals > 15000] = np.nan
pixcvec_latvals[pixcvec_latvals == 0] = np.nan
pixcvec_lonvals[pixcvec_lonvals == 0] = np.nanplt.scatter(x=pixcvec_lonvals, y=pixcvec_latvals, c=pixcvec_htvals)
plt.colorbar().set_label('Height (m)')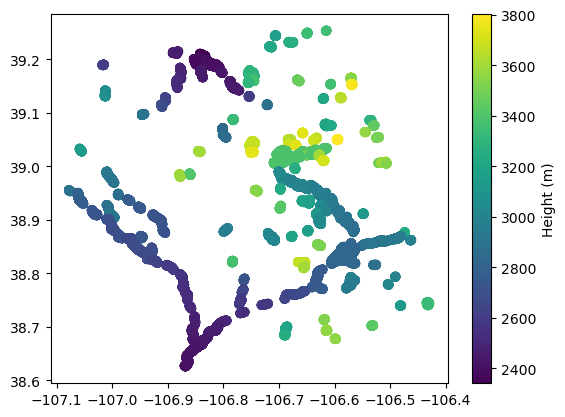
5. Raster NetCDF
Search for data of interest
raster_results = earthaccess.search_data(short_name = 'SWOT_L2_HR_Raster_2.0',
temporal = ('2024-01-25 00:00:00', '2024-07-15 23:59:59'), # can also specify by time
granule_name = '*100m*', # here we filter by files with '100m' in the name (This collection has two resolution options: 100m & 250m)
bounding_box = (-106.62, 38.809, -106.54, 38.859)) # Lake Travis near Austin, TXGranules found: 83Let’s download one data file.
earthaccess.download([raster_results[0]], "./data_downloads") Getting 1 granules, approx download size: 0.05 GB['data_downloads\\SWOT_L2_HR_Raster_100m_UTM13S_N_x_x_x_010_106_043F_20240128T183530_20240128T183551_PIC0_01.nc']Open data with xarray
First, we’ll programmatically get the filename we just downloaded and then view the file via xarray.
ds_raster = xr.open_mfdataset(f'data_downloads/SWOT_L2_HR_Raster*', engine='h5netcdf')
ds_raster<xarray.Dataset>
Dimensions: (x: 1520, y: 1519)
Coordinates:
* x (x) float64 2.969e+05 2.97e+05 ... 4.488e+05
* y (y) float64 4.274e+06 4.274e+06 ... 4.426e+06
Data variables: (12/39)
crs object ...
longitude (y, x) float64 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
latitude (y, x) float64 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
wse (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
wse_qual (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
wse_qual_bitwise (y, x) float64 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
... ...
load_tide_fes (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
load_tide_got (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
pole_tide (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
model_dry_tropo_cor (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
model_wet_tropo_cor (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
iono_cor_gim_ka (y, x) float32 dask.array<chunksize=(1519, 1520), meta=np.ndarray>
Attributes: (12/49)
Conventions: CF-1.7
title: Level 2 KaRIn High Rate Raster Data Product
source: Ka-band radar interferometer
history: 2024-02-02T15:24:17Z : Creation
platform: SWOT
references: V1.2.1
... ...
x_min: 296900.0
x_max: 448800.0
y_min: 4274000.0
y_max: 4425800.0
institution: CNES
product_version: 01Quick interactive plot with hvplot
Note: this is not filtered by quality
ds_raster.wse.hvplot.image(y='y', x='x')
