import xarray as xr
xr.set_options(keep_attrs=True)
import hvplot.xarrayXarray
imported on: 2026-02-13
This notebook is from NASA Openscapes 2021 Cloud Hackathon Repository
The original source for this document is https://github.com/NASA-Openscapes/2021-Cloud-Hackathon/blob/main/tutorials/03_Xarray_hvplot.ipynb
03. Introduction to xarray
Why do we need xarray?
As Geoscientists, we often work with time series of data with two or more dimensions: a time series of calibrated, orthorectified satellite images; two-dimensional grids of surface air temperature from an atmospheric reanalysis; or three-dimensional (level, x, y) cubes of ocean salinity from an ocean model. These data are often provided in GeoTIFF, NetCDF or HDF format with rich and useful metadata that we want to retain, or even use in our analysis. Common analyses include calculating means, standard deviations and anomalies over time or one or more spatial dimensions (e.g. zonal means). Model output often includes multiple variables that you want to apply similar analyses to.

The schematic above shows a typical data structure for multi-dimensional data. There are two data cubes, one for temperature and one for precipitation. Common coordinate variables, in this case latitude, longitude and time are associated with each variable. Each variable, including coordinate variables, will have a set of attributes: name, units, missing value, etc. The file containing the data may also have attributes: source of the data, model name coordinate reference system if the data are projected. Writing code using low-level packages such as netcdf4 and numpy to read the data, then perform analysis, and write the results to file is time consuming and prone to errors.
What is xarray
xarray is an open-source project and python package to work with labelled multi-dimensional arrays. It is leverages numpy, pandas, matplotlib and dask to build Dataset and DataArray objects with built-in methods to subset, analyze, interpolate, and plot multi-dimensional data. It makes working with multi-dimensional data cubes efficient and fun. It will change your life for the better. You’ll be more attractive, more interesting, and better equiped to take on lifes challenges.
What you will learn from this tutorial
In this tutorial you will learn how to:
- load a netcdf file into
xarray - interrogate the
Datasetand understand the difference betweenDataArrayandDataset - subset a
Dataset - calculate annual and monthly mean fields
- calculate a time series of zonal means
- plot these results
As always, we’ll start by importing xarray. We’ll follow convention by giving the module the shortname xr
I’m going to use one of xarray’s tutorial datasets. In this case, air temperature from the NCEP reanalysis. I’ll assign the result of the open_dataset to ds. I may change this to access a dataset directly
ds = xr.tutorial.open_dataset("air_temperature")As we are in an interactive environment, we can just type ds to see what we have.
ds<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 ...
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...- lat: 25
- time: 2920
- lon: 53
- lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- air(time, lat, lon)float32...
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
[3869000 values with dtype=float32]
- Conventions :
- COARDS
- title :
- 4x daily NMC reanalysis (1948)
- description :
- Data is from NMC initialized reanalysis (4x/day). These are the 0.9950 sigma level values.
- platform :
- Model
- references :
- http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanalysis.html
First thing to notice is that ds is an xarray.Dataset object. It has dimensions, lat, lon, and time. It also has coordinate variables with the same names as these dimensions. These coordinate variables are 1-dimensional. This is a NetCDF convention. The Dataset contains one data variable, air. This has dimensions (time, lat, lon).
Clicking on the document icon reveals attributes for each variable. Clicking on the disk icon reveals a representation of the data.
Each of the data and coordinate variables can be accessed and examined using the variable name as a key.
ds.air<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
[3869000 values with dtype=float32]
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]- time: 2920
- lat: 25
- lon: 53
- ...
[3869000 values with dtype=float32]
- lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
ds['air']<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
[3869000 values with dtype=float32]
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]- time: 2920
- lat: 25
- lon: 53
- ...
[3869000 values with dtype=float32]
- lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
These are xarray.DataArray objects. This is the basic building block for xarray.
Variables can also be accessed as attributes of ds.
ds.time<xarray.DataArray 'time' (time: 2920)>
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000',
'2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000',
'2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
standard_name: time
long_name: Time- time: 2920
- 2013-01-01 2013-01-01T06:00:00 ... 2014-12-31T18:00:00
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]') - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- standard_name :
- time
- long_name :
- Time
A major difference between accessing a variable as an attribute versus using a key is that the attribute is read-only but the key method can be used to update the variable. For example, if I want to convert the units of air from Kelvin to degrees Celsius.
ds['air'] = ds.air - 273.15This approach can also be used to add new variables
ds['air_kelvin'] = ds.air + 273.15It is helpful to update attributes such as units, this saves time, confusion and mistakes, especially when you save the dataset.
ds['air'].attrs['units'] = 'degC'ds<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 -31.95 -30.65 -29.65 ... 23.04 22.54
air_kelvin (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...- lat: 25
- time: 2920
- lon: 53
- lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- air(time, lat, lon)float32-31.95 -30.65 ... 23.04 22.54
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([[[-31.949997, -30.649994, -29.649994, ..., -40.350006, -37.649994, -34.550003], [-29.350006, -28.649994, -28.449997, ..., -40.350006, -37.850006, -33.850006], [-23.149994, -23.350006, -24.259995, ..., -39.949997, -36.759995, -31.449997], ..., [ 23.450012, 23.049988, 23.25 , ..., 22.25 , 21.950012, 21.549988], [ 22.75 , 23.049988, 23.640015, ..., 22.75 , 22.75 , 22.049988], [ 23.140015, 23.640015, 23.950012, ..., 23.75 , 23.640015, 23.450012]], [[-31.050003, -30.449997, -30.050003, ..., -41.149994, -39.550003, -37.350006], [-29.550003, -29.050003, -28.949997, ..., -42.149994, -40.649994, -37.449997], [-19.949997, -20.259995, -21.050003, ..., -42.350006, -39.759995, -34.649994], ... [ 20.540009, 20.73999 , 22.23999 , ..., 21.940002, 21.540009, 21.140015], [ 23.140015, 24.040009, 24.440002, ..., 22.140015, 21.940002, 21.23999 ], [ 24.640015, 25.23999 , 25.339996, ..., 22.540009, 22.339996, 22.040009]], [[-28.059998, -28.86 , -29.86 , ..., -31.460007, -31.660004, -31.36 ], [-23.259995, -23.86 , -24.759995, ..., -33.559998, -32.86 , -31.460007], [-10.160004, -10.959991, -11.76001 , ..., -33.259995, -30.559998, -26.86 ], ..., [ 20.640015, 20.540009, 21.940002, ..., 22.140015, 21.940002, 21.540009], [ 22.940002, 23.73999 , 24.040009, ..., 22.540009, 22.540009, 22.040009], [ 24.540009, 24.940002, 24.940002, ..., 23.339996, 23.040009, 22.540009]]], dtype=float32) - air_kelvin(time, lat, lon)float32241.2 242.5 243.5 ... 296.2 295.7
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 , 238.59999], [243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999, 239.29999], [250. , 249.79999, 248.89 , ..., 233.2 , 236.39 , 241.7 ], ..., [296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 , 294.69998], [295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 , 295.19998], [296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 , 296.6 ]], [[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999, 235.79999], [243.59999, 244.09999, 244.2 , ..., 231. , 232.5 , 235.7 ], [253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 , 238.5 ], ... [293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 , 294.29 ], [296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 , 294.38998], [297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 , 295.19 ]], [[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999, 241.79 ], [249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 , 241.68999], [262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 , 246.29 ], ..., [293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 , 294.69 ], [296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 , 295.19 ], [297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 , 295.69 ]]], dtype=float32)
- Conventions :
- COARDS
- title :
- 4x daily NMC reanalysis (1948)
- description :
- Data is from NMC initialized reanalysis (4x/day). These are the 0.9950 sigma level values.
- platform :
- Model
- references :
- http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanalysis.html
Subsetting and Indexing
Subsetting and indexing methods depend on whether you are working with a Dataset or DataArray. A DataArray can be accessed using positional indexing just like a numpy array. To access the temperature field for the first time step, you do the following.
ds['air'][0,:,:]<xarray.DataArray 'air' (lat: 25, lon: 53)>
array([[-31.949997, -30.649994, -29.649994, ..., -40.350006, -37.649994,
-34.550003],
[-29.350006, -28.649994, -28.449997, ..., -40.350006, -37.850006,
-33.850006],
[-23.149994, -23.350006, -24.259995, ..., -39.949997, -36.759995,
-31.449997],
...,
[ 23.450012, 23.049988, 23.25 , ..., 22.25 , 21.950012,
21.549988],
[ 22.75 , 23.049988, 23.640015, ..., 22.75 , 22.75 ,
22.049988],
[ 23.140015, 23.640015, 23.950012, ..., 23.75 , 23.640015,
23.450012]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
time datetime64[ns] 2013-01-01
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degC
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]- lat: 25
- lon: 53
- -31.95 -30.65 -29.65 -29.15 -29.05 ... 24.64 24.45 23.75 23.64 23.45
array([[-31.949997, -30.649994, -29.649994, ..., -40.350006, -37.649994, -34.550003], [-29.350006, -28.649994, -28.449997, ..., -40.350006, -37.850006, -33.850006], [-23.149994, -23.350006, -24.259995, ..., -39.949997, -36.759995, -31.449997], ..., [ 23.450012, 23.049988, 23.25 , ..., 22.25 , 21.950012, 21.549988], [ 22.75 , 23.049988, 23.640015, ..., 22.75 , 22.75 , 22.049988], [ 23.140015, 23.640015, 23.950012, ..., 23.75 , 23.640015, 23.450012]], dtype=float32) - lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time()datetime64[ns]2013-01-01
- standard_name :
- time
- long_name :
- Time
array('2013-01-01T00:00:00.000000000', dtype='datetime64[ns]')
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
Note this returns a DataArray with coordinates but not attributes.
However, the real power is being able to access variables using coordinate variables. I can get the same subset using the following. (It’s also more explicit about what is being selected and robust in case I modify the DataArray and expect the same output.)
ds['air'].sel(time='2013-01-01').time<xarray.DataArray 'time' (time: 4)>
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000',
'2013-01-01T12:00:00.000000000', '2013-01-01T18:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 2013-01-01 ... 2013-01-01T18:00:00
Attributes:
standard_name: time
long_name: Time- time: 4
- 2013-01-01 2013-01-01T06:00:00 2013-01-01T12:00:00 2013-01-01T18:00:00
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', '2013-01-01T18:00:00.000000000'], dtype='datetime64[ns]') - time(time)datetime64[ns]2013-01-01 ... 2013-01-01T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', '2013-01-01T18:00:00.000000000'], dtype='datetime64[ns]')
- standard_name :
- time
- long_name :
- Time
ds.air.sel(time='2013-01-01')<xarray.DataArray 'air' (time: 4, lat: 25, lon: 53)>
array([[[-31.949997, -30.649994, -29.649994, ..., -40.350006,
-37.649994, -34.550003],
[-29.350006, -28.649994, -28.449997, ..., -40.350006,
-37.850006, -33.850006],
[-23.149994, -23.350006, -24.259995, ..., -39.949997,
-36.759995, -31.449997],
...,
[ 23.450012, 23.049988, 23.25 , ..., 22.25 ,
21.950012, 21.549988],
[ 22.75 , 23.049988, 23.640015, ..., 22.75 ,
22.75 , 22.049988],
[ 23.140015, 23.640015, 23.950012, ..., 23.75 ,
23.640015, 23.450012]],
[[-31.050003, -30.449997, -30.050003, ..., -41.149994,
-39.550003, -37.350006],
[-29.550003, -29.050003, -28.949997, ..., -42.149994,
-40.649994, -37.449997],
[-19.949997, -20.259995, -21.050003, ..., -42.350006,
-39.759995, -34.649994],
...
[ 22.450012, 22.25 , 22.25 , ..., 23.140015,
22.140015, 21.850006],
[ 23.049988, 23.350006, 23.140015, ..., 23.25 ,
22.850006, 22.450012],
[ 23.25 , 23.140015, 23.25 , ..., 23.850006,
23.850006, 23.640015]],
[[-31.259995, -31.350006, -31.350006, ..., -38.759995,
-37.649994, -35.550003],
[-26.850006, -27.850006, -28.949997, ..., -42.259995,
-41.649994, -38.649994],
[-16.549988, -18.449997, -21.050003, ..., -42.449997,
-41.350006, -37.050003],
...,
[ 23.450012, 23.25 , 22.850006, ..., 23.350006,
22.640015, 22.140015],
[ 23.850006, 24.350006, 23.950012, ..., 23.640015,
23.450012, 23.140015],
[ 24.350006, 24.549988, 24.350006, ..., 24.640015,
24.850006, 24.75 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2013-01-01T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degC
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]- time: 4
- lat: 25
- lon: 53
- -31.95 -30.65 -29.65 -29.15 -29.05 ... 25.45 25.05 24.64 24.85 24.75
array([[[-31.949997, -30.649994, -29.649994, ..., -40.350006, -37.649994, -34.550003], [-29.350006, -28.649994, -28.449997, ..., -40.350006, -37.850006, -33.850006], [-23.149994, -23.350006, -24.259995, ..., -39.949997, -36.759995, -31.449997], ..., [ 23.450012, 23.049988, 23.25 , ..., 22.25 , 21.950012, 21.549988], [ 22.75 , 23.049988, 23.640015, ..., 22.75 , 22.75 , 22.049988], [ 23.140015, 23.640015, 23.950012, ..., 23.75 , 23.640015, 23.450012]], [[-31.050003, -30.449997, -30.050003, ..., -41.149994, -39.550003, -37.350006], [-29.550003, -29.050003, -28.949997, ..., -42.149994, -40.649994, -37.449997], [-19.949997, -20.259995, -21.050003, ..., -42.350006, -39.759995, -34.649994], ... [ 22.450012, 22.25 , 22.25 , ..., 23.140015, 22.140015, 21.850006], [ 23.049988, 23.350006, 23.140015, ..., 23.25 , 22.850006, 22.450012], [ 23.25 , 23.140015, 23.25 , ..., 23.850006, 23.850006, 23.640015]], [[-31.259995, -31.350006, -31.350006, ..., -38.759995, -37.649994, -35.550003], [-26.850006, -27.850006, -28.949997, ..., -42.259995, -41.649994, -38.649994], [-16.549988, -18.449997, -21.050003, ..., -42.449997, -41.350006, -37.050003], ..., [ 23.450012, 23.25 , 22.850006, ..., 23.350006, 22.640015, 22.140015], [ 23.850006, 24.350006, 23.950012, ..., 23.640015, 23.450012, 23.140015], [ 24.350006, 24.549988, 24.350006, ..., 24.640015, 24.850006, 24.75 ]]], dtype=float32) - lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time(time)datetime64[ns]2013-01-01 ... 2013-01-01T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', '2013-01-01T18:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
I can also do slices. I’ll extract temperatures for the state of Colorado. The bounding box for the state is [-109 E, -102 E, 37 N, 41 N].
In the code below, pay attention to both the order of the coordinates and the range of values. The first value of the lat coordinate variable is 41 N, the second value is 37 N. Unfortunately, xarray expects slices of coordinates to be in the same order as the coordinates. Note lon is 0 to 360 not -180 to 180, and I let python calculate it for me within the slice.
ds.air.sel(lat=slice(41.,37.), lon=slice(360-109,360-102))<xarray.DataArray 'air' (time: 2920, lat: 2, lon: 3)>
array([[[-10.049988 , -9.25 , -8.75 ],
[ -6.25 , -6.549988 , -6.3599854]],
[[-18.149994 , -14.950012 , -9.950012 ],
[-13.649994 , -11.049988 , -7.25 ]],
[[-20.449997 , -18.649994 , -13.359985 ],
[-19.350006 , -16.950012 , -11.25 ]],
...,
[[-24.460007 , -28.259995 , -25.759995 ],
[-16.959991 , -24.059998 , -24.059998 ]],
[[-24.36 , -26.160004 , -23.460007 ],
[-15.959991 , -22.86 , -22.960007 ]],
[[-17.559998 , -15.359985 , -13.660004 ],
[-13.76001 , -15.959991 , -14.459991 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 40.0 37.5
* lon (lon) float32 252.5 255.0 257.5
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degC
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]- time: 2920
- lat: 2
- lon: 3
- -10.05 -9.25 -8.75 -6.25 -6.55 ... -15.36 -13.66 -13.76 -15.96 -14.46
array([[[-10.049988 , -9.25 , -8.75 ], [ -6.25 , -6.549988 , -6.3599854]], [[-18.149994 , -14.950012 , -9.950012 ], [-13.649994 , -11.049988 , -7.25 ]], [[-20.449997 , -18.649994 , -13.359985 ], [-19.350006 , -16.950012 , -11.25 ]], ..., [[-24.460007 , -28.259995 , -25.759995 ], [-16.959991 , -24.059998 , -24.059998 ]], [[-24.36 , -26.160004 , -23.460007 ], [-15.959991 , -22.86 , -22.960007 ]], [[-17.559998 , -15.359985 , -13.660004 ], [-13.76001 , -15.959991 , -14.459991 ]]], dtype=float32) - lat(lat)float3240.0 37.5
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([40. , 37.5], dtype=float32)
- lon(lon)float32252.5 255.0 257.5
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([252.5, 255. , 257.5], dtype=float32)
- time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
What if we want temperature for a point, for example Denver, CO (39.72510678889283 N, -104.98785545855408 E). xarray can handle this! If we just want data from the nearest grid point, we can use sel and specify the method as “nearest”.
denver_lat, denver_lon = 39.72510678889283, -104.98785545855408ds.air.sel(lat=denver_lat, lon=360+denver_lon, method='nearest').hvplot()If we want to interpolate, we can use interp(). In this case I use linear or bilinear interpolation.
interp() can also be used to resample data to a new grid and even reproject data
ds.air.interp(lat=denver_lat, lon=360+denver_lon, method='linear')<xarray.DataArray 'air' (time: 2920)>
array([ -8.95085077, -14.49752791, -18.43715163, ..., -27.78736503,
-25.78552388, -15.41780902])
Coordinates:
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
lat float64 39.73
lon float64 255.0
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degC
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]- time: 2920
- -8.951 -14.5 -18.44 -11.33 -8.942 ... -22.4 -27.79 -25.79 -15.42
array([ -8.95085077, -14.49752791, -18.43715163, ..., -27.78736503, -25.78552388, -15.41780902]) - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]') - lat()float6439.73
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array(39.72510679)
- lon()float64255.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array(255.01214454)
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
sel() and interp() can also be used on Dataset objects.
ds.sel(lat=slice(41,37), lon=slice(360-109,360-102))<xarray.Dataset>
Dimensions: (lat: 2, time: 2920, lon: 3)
Coordinates:
* lat (lat) float32 40.0 37.5
* lon (lon) float32 252.5 255.0 257.5
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 -10.05 -9.25 -8.75 ... -15.96 -14.46
air_kelvin (time, lat, lon) float32 263.1 263.9 264.4 ... 259.4 257.2 258.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...- lat: 2
- time: 2920
- lon: 3
- lat(lat)float3240.0 37.5
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([40. , 37.5], dtype=float32)
- lon(lon)float32252.5 255.0 257.5
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([252.5, 255. , 257.5], dtype=float32)
- time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- air(time, lat, lon)float32-10.05 -9.25 ... -15.96 -14.46
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([[[-10.049988 , -9.25 , -8.75 ], [ -6.25 , -6.549988 , -6.3599854]], [[-18.149994 , -14.950012 , -9.950012 ], [-13.649994 , -11.049988 , -7.25 ]], [[-20.449997 , -18.649994 , -13.359985 ], [-19.350006 , -16.950012 , -11.25 ]], ..., [[-24.460007 , -28.259995 , -25.759995 ], [-16.959991 , -24.059998 , -24.059998 ]], [[-24.36 , -26.160004 , -23.460007 ], [-15.959991 , -22.86 , -22.960007 ]], [[-17.559998 , -15.359985 , -13.660004 ], [-13.76001 , -15.959991 , -14.459991 ]]], dtype=float32) - air_kelvin(time, lat, lon)float32263.1 263.9 264.4 ... 257.2 258.7
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([[[263.1 , 263.9 , 264.4 ], [266.9 , 266.6 , 266.79 ]], [[255. , 258.19998, 263.19998], [259.5 , 262.1 , 265.9 ]], [[252.7 , 254.5 , 259.79 ], [253.79999, 256.19998, 261.9 ]], ..., [[248.68999, 244.89 , 247.39 ], [256.19 , 249.09 , 249.09 ]], [[248.79 , 246.98999, 249.68999], [257.19 , 250.29 , 250.18999]], [[255.59 , 257.79 , 259.49 ], [259.38998, 257.19 , 258.69 ]]], dtype=float32)
- Conventions :
- COARDS
- title :
- 4x daily NMC reanalysis (1948)
- description :
- Data is from NMC initialized reanalysis (4x/day). These are the 0.9950 sigma level values.
- platform :
- Model
- references :
- http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanalysis.html
ds.interp(lat=denver_lat, lon=360+denver_lon, method='linear')<xarray.Dataset>
Dimensions: (time: 2920)
Coordinates:
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
lat float64 39.73
lon float64 255.0
Data variables:
air (time) float64 -8.951 -14.5 -18.44 ... -27.79 -25.79 -15.42
air_kelvin (time) float64 264.2 258.7 254.7 261.8 ... 245.4 247.4 257.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...- time: 2920
- time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]') - lat()float6439.73
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array(39.72510679)
- lon()float64255.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array(255.01214454)
- air(time)float64-8.951 -14.5 ... -25.79 -15.42
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([ -8.95085077, -14.49752791, -18.43715163, ..., -27.78736503, -25.78552388, -15.41780902]) - air_kelvin(time)float64264.2 258.7 254.7 ... 247.4 257.7
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([264.19914312, 258.65246598, 254.71284227, ..., 245.36262886, 247.36447002, 257.73218487])
- Conventions :
- COARDS
- title :
- 4x daily NMC reanalysis (1948)
- description :
- Data is from NMC initialized reanalysis (4x/day). These are the 0.9950 sigma level values.
- platform :
- Model
- references :
- http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanalysis.html
Analysis
As a simple example, let’s try to calculate a mean field for the whole time range.
ds.mean(dim='time').hvplot()We can also calculate a zonal mean (averaging over longitude)
ds.mean(dim='lon').hvplot()Other aggregation methods include min(), max(), std(), along with others.
ds.std(dim='time').hvplot()The data we have are in 6h timesteps. This can be resampled to daily or monthly. If you are familiar with pandas, xarray uses the same methods.
ds.resample(time='M').mean().hvplot()ds_mon = ds.resample(time='M').mean()
ds_mon<xarray.Dataset>
Dimensions: (time: 24, lat: 25, lon: 53)
Coordinates:
* time (time) datetime64[ns] 2013-01-31 2013-02-28 ... 2014-12-31
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
Data variables:
air (time, lat, lon) float32 -28.68 -28.49 -28.48 ... 24.57 24.56
air_kelvin (time, lat, lon) float32 244.5 244.7 244.7 ... 297.7 297.7 297.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...- time: 24
- lat: 25
- lon: 53
- time(time)datetime64[ns]2013-01-31 ... 2014-12-31
array(['2013-01-31T00:00:00.000000000', '2013-02-28T00:00:00.000000000', '2013-03-31T00:00:00.000000000', '2013-04-30T00:00:00.000000000', '2013-05-31T00:00:00.000000000', '2013-06-30T00:00:00.000000000', '2013-07-31T00:00:00.000000000', '2013-08-31T00:00:00.000000000', '2013-09-30T00:00:00.000000000', '2013-10-31T00:00:00.000000000', '2013-11-30T00:00:00.000000000', '2013-12-31T00:00:00.000000000', '2014-01-31T00:00:00.000000000', '2014-02-28T00:00:00.000000000', '2014-03-31T00:00:00.000000000', '2014-04-30T00:00:00.000000000', '2014-05-31T00:00:00.000000000', '2014-06-30T00:00:00.000000000', '2014-07-31T00:00:00.000000000', '2014-08-31T00:00:00.000000000', '2014-09-30T00:00:00.000000000', '2014-10-31T00:00:00.000000000', '2014-11-30T00:00:00.000000000', '2014-12-31T00:00:00.000000000'], dtype='datetime64[ns]') - lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32)
- air(time, lat, lon)float32-28.68 -28.49 ... 24.57 24.56
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degC
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([[[-28.68323 , -28.486452 , -28.479755 , ..., -30.658554 , -29.743628 , -28.474194 ], [-26.076784 , -26.127504 , -26.4225 , ..., -32.5679 , -31.105167 , -28.442825 ], [-22.770565 , -23.31516 , -24.042498 , ..., -31.165657 , -28.38291 , -24.144924 ], ..., [ 22.688152 , 22.00097 , 21.773153 , ..., 22.218397 , 21.734531 , 21.118395 ], [ 23.31952 , 23.16702 , 22.698233 , ..., 22.43775 , 22.190727 , 21.715578 ], [ 23.903486 , 23.89203 , 23.585333 , ..., 23.154608 , 22.947426 , 22.889124 ]], [[-32.41607 , -32.44866 , -32.738483 , ..., -31.54482 , -30.430185 , -29.205448 ], [-31.216885 , -31.08063 , -31.236965 , ..., -32.135708 , -30.825186 , -28.42241 ], [-27.826433 , -28.123934 , -28.78045 , ..., -29.734114 , -27.383936 , -23.491434 ], ... [ 24.899088 , 24.200085 , 24.072004 , ..., 24.861843 , 24.510258 , 23.995668 ], [ 25.815008 , 25.661922 , 25.121607 , ..., 24.954088 , 25.071083 , 24.735588 ], [ 26.023424 , 26.06767 , 25.74576 , ..., 25.566338 , 25.591848 , 25.630259 ]], [[-26.348473 , -26.260897 , -26.380894 , ..., -33.07903 , -32.067986 , -30.868315 ], [-25.419994 , -24.849277 , -24.405483 , ..., -34.531376 , -32.82783 , -30.179682 ], [-23.181051 , -23.56476 , -23.574757 , ..., -35.446938 , -31.91259 , -26.923311 ], ..., [ 23.299198 , 22.541454 , 22.60839 , ..., 23.378307 , 23.067505 , 22.662996 ], [ 24.295895 , 24.286139 , 24.031782 , ..., 23.80259 , 23.908312 , 23.579037 ], [ 24.897346 , 25.076134 , 24.909689 , ..., 24.547583 , 24.573233 , 24.560413 ]]], dtype=float32) - air_kelvin(time, lat, lon)float32244.5 244.7 244.7 ... 297.7 297.7
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
array([[[244.4667 , 244.66354, 244.67027, ..., 242.49142, 243.40633, 244.67577], [247.07323, 247.02248, 246.7275 , ..., 240.58205, 242.04489, 244.70726], [250.37941, 249.83484, 249.10748, ..., 241.98434, 244.76712, 249.00505], ..., [295.83795, 295.15085, 294.9229 , ..., 295.36826, 294.88437, 294.26828], [296.46942, 296.31686, 295.84802, ..., 295.5876 , 295.34058, 294.86536], [297.05316, 297.0418 , 296.73517, ..., 296.30438, 296.09732, 296.0389 ]], [[240.73384, 240.7013 , 240.4115 , ..., 241.60518, 242.71988, 243.94455], [241.93309, 242.06935, 241.913 , ..., 241.01428, 242.32481, 244.72758], [245.32361, 245.0261 , 244.36955, ..., 243.41588, 245.7661 , 249.65858], ... [298.04895, 297.35007, 297.22195, ..., 298.01172, 297.66013, 297.14554], [298.96484, 298.81186, 298.27136, ..., 298.10403, 298.22104, 297.88547], [299.17334, 299.2175 , 298.89566, ..., 298.71625, 298.74167, 298.7802 ]], [[246.80156, 246.88907, 246.76907, ..., 240.07089, 241.08206, 242.2817 ], [247.72998, 248.30064, 248.74443, ..., 238.61859, 240.3222 , 242.97026], [249.96893, 249.58516, 249.57521, ..., 237.70308, 241.23743, 246.22667], ..., [296.4491 , 295.6914 , 295.75824, ..., 296.52817, 296.21747, 295.8128 ], [297.44586, 297.43613, 297.1817 , ..., 296.95242, 297.05823, 296.72897], [298.0472 , 298.22598, 298.0595 , ..., 297.6975 , 297.72318, 297.71024]]], dtype=float32)
- Conventions :
- COARDS
- title :
- 4x daily NMC reanalysis (1948)
- description :
- Data is from NMC initialized reanalysis (4x/day). These are the 0.9950 sigma level values.
- platform :
- Model
- references :
- http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanalysis.html
This is a really short time series but as an example, let’s calculate a monthly climatology (at least for 2 months). For this we can use groupby()
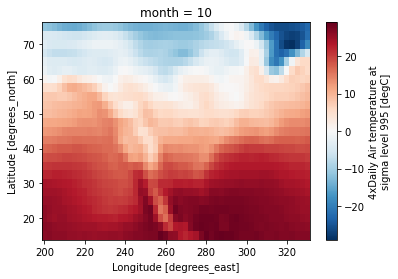
ds_clim = ds_mon.groupby(ds_mon.time.dt.month).mean()Plot results
Finally, let’s plot the results! This will plot the lat/lon axes of the original ds DataArray.
ds_clim.air.sel(month=10).plot()